Microbiology often has been defined as the study of organisms and agents too small to be seen clearly by the unaided eye—that is, the study of microorganisms. Because objects less than about one millimeter in diameter cannot be seen clearly and must be examined with a microscope, microbiology is concerned primarily with organisms and agents this small and smaller.
Microbial World
Microorganisms are everywhere. Almost every natural surface is colonized by microbes (including our skin). Some microorganisms can live quite happily in boiling hot springs, whereas others form complex microbial communities in frozen sea ice.
Most microorganisms are harmless to humans. You swallow millions of microbes every day with no ill effects. In fact, we are dependent on microbes to help us digest our food and to protect our bodies from pathogens. Microbes also keep the biosphere running by carrying out essential functions such as decomposition of dead animals and plants.
Microbes are the dominant form of life on planet Earth. More than half the biomass on Earth consists of microorganisms, whereas animals constitute only 15% of the mass of living organisms on Earth.
This Microbiology course deals with
- How and where they live
- Their structure
- How they derive food and energy
- Functions of soil micro flora
- Role in nutrient transformation
- Relation with plant
- Importance in Industries
The microorganisms can be divided into two distinct groups based on the nucleus structure:
Prokaryotes – The organism lacking true nucleus (membrane enclosed chromosome and nucleolus) and other organelles like mitochondria, golgi body, entoplasmic reticulum etc. are referred as Prokaryotes. (Ex : Bacteria, archaea)
Eukaryotes – The organism possessing membrane enclosed nucleus and other cell organelles are referred as Eukaryotes (Ex : algae, fungi, protozoa)
The microorganisms were divided into 6 distinct groups based on the phylogenic, morphological and physiological characters.
The major groups of microorganisms are
- Bacteria are phylogenetically related group of unicellular prokaryotic organisms distinct from archeae
- Archaea is phylogenetically related group of prokaryotes which are primitive and distinct from bacteria
- Fungi are group of eukaryotic organisms lacking chlorophyll. They range in size and shape from single celled yeast to multicellular mushrooms.
- Algae refer the group of eukaryotic organisms with chlorophyll. They range in size and shape from single celled algae (Ex: Chlorella) to complex cellular structured plant like algae (Ex. Kelp)
- Protozoa are group of eukaryotic organisms lack of cell wall. The morphology, nutrition and physiology is different from other groups
- Viruses are group of non-cellular organisms, parasite or pathogen to plant, animals and other microorganisms. They are too small and cab be visualized only under electron microscopes
History of Microbiology in brief
Obviously human have had to deal with microbes even before the recorded history. The first record of human using comes from ancient tablets from mid east.
Babylonians were using yeast to make beer over 8000 years ago and acetic acid bacteria to make vinegar over 6000 years ago.
About 5000 years ago, Persia (Now Iran) region recorded the wine making.
The Romans had God for that were specific for microorganisms. The roman God of mold and mildew was “Robigus” and “Robigo” which means crop rust. (Rust is one of the plant disease caused by fungus). God Robigus was very much feared because of crop lost.
About 2000 years ago, Romans proposed that diseases were caused by tiny animals. But, fundamentalist religions had a strong hold over the progress.
The real microbiology history starts from 1600s, when people began to make crude lenses and microscopes.
HIGHLIGHTS IN THE HISTORY OF MICROBIOLOGY
Effects of Disease on Civilization
- Infectious diseases have played major roles in shaping human history
- Bubonic Plague epidemic of mid 1300’s, the “Great Plague”, reduced population of western Europe by 25%. Plague bacterium was carried by fleas, spread from China via trade routes and poor hygiene. As fleas became established in rat populations in Western Europe, disease became major crisis.
- Smallpox and other infectious diseases introduced by European explorers to the Americas in 1500’s were responsible for decimating Native American populations. Example: In the century after Hernan Cortez’s arrival in Mexico, the Aztec population declined from about 20 million to about 1.6 million, mainly because of disease.
- Infectious diseases have killed more soldiers than battles in all wars up to WW II. Example: in U. S. Civil war, 93,000 Union soldiers died in direct combat; 210,000 died as a result of infections.
- Until late 1800’s, no one had proved that infectious diseases were caused by specific microbes, so the possibility of prevention or treatment had no sound empirical base.

Brueghel: The Triumph of Death (1560)
Discovery of Microbes
- To see microbes, you need a microscope. The first microscope was invented by Antony van Leeuwenhoek (1632-1723), a Dutch businessman.
- Leeuwenhoek took up lens grinding to make magnifiying glasses so he could examine fine weave of fabrics. In testing his lenses, he discovered many small creatures he called “animalcules” in samples such as pond water. His best lenses could magnify 300-500X.
- Leeuwenhoek microscopes were crude, relied on a single lens held in a metal plate.
- Leeuwenhoek described many previously unseen life forms, including different forms of bacteria, mold spores, etc. Leeuwenhoek reported discoveries to Royal Society from 1670’s on, firmly established existence of microbes. Nevertheless, the significance of this discovery was not apparent for almost 200 years.
Antony van Leeuwenhoek.
Origin of Life Controversy
- Where did microbes come from? Many believed they arose from simple materials by process of spontaneous generation. This notion had been posited by Aristotle (382-322 B.C.) and other Greek philosophers to explain decay and appearance of animals such as flies and frogs, and was widely held as common sense even in 1700’s and 1800’s.
- Francisco Redi (1626-1697) demonstrated that flies did not arise spontaneously from rotting meat by simple experiment. If jar of meat was covered by fine muslin, maggots did not arise. However, the simpler life forms discovered by Leeuwenhoek lacked visible complexity, and most people still believed these could arise spontaneously.


- John Needham (1731-1781), a Scottish clergyman and naturalist, showed that mirobes grew in soups exposed to air. Claimed existence of a “life force” present in inorganic matter that could cause spontaneous generation. One of his more convincing demonstrations was to boil some soup (briefly), pour into clean flasks
 with cork lids, and show that microbes would soon arise.
with cork lids, and show that microbes would soon arise.
- Lazzaro Spallanzani (1729-1799) claimed Needham’s organisms came from heat-resistant microbes. If flasks were boiled long enough (1-2 h), nothing grew. But Needham countered that prolonged heating destroyed the “life force”.

- Louis Pasteur (1822-1895) was passionate believer that life only originated from previous life, developed several experiments that finally deflated claims for spontaneous generation. Pasteur filtered air through cotton to trap airborne materials, then dissolved the cotton and examined the particulate matter under a microscope; many bacteria and spores of other life forms such as molds were present. Since most skeptics kept arguing that overheating killed the life force present in air, Pasteur developed and ingenious experiment using a swan neck flask that allowed fresh air to remain in contact with boiled materials. The long passageway prevented airborne microbes from reaching the nutrient liquid, without impeding access to air. One of Pasteur’s flasks is still sterile after 100+ years of being exposed to the air (Pasteur Institute, Paris).

Louis Pasteur
Spontaneous Generation theory
From earliest times, people had believed in spontaneous generation—that living organisms could develop from nonliving matter. Even the great Aristotle (384–322 B.C.) thought some of the simpler invertebrates could arise by spontaneous generation. This view finally was challenged by the Italian physician Francesco Redi (1626–1697), who carried out a series of experiments on decaying meat and its ability to produce maggots spontaneously. Redi placed meat in three containers. One was uncovered, a second was covered with paper, and the third was covered with a fine gauze that would exclude flies. Flies laid their eggs on the uncovered meat and maggots developed. The other two pieces of meat did not produce maggots spontaneously. However, flies were attracted to the gauze-covered container and laid their eggs on the gauze; these eggs produced maggots. Thus the generation of maggots by decaying meat resulted from the presence of fly eggs, and meat did not spontaneously generate maggots as previously believed. Similar experiments by others helped discredit the theory for larger organisms.
Leeuwenhoek’s discovery of microorganisms renewed the controversy. Some proposed that microorganisms arose by spontaneous generation even though larger organisms did not. They pointed out that boiled extracts of hay or meat would give rise to microorganisms after sitting for a while. In 1748 the English priest John Needham (1713–1781) reported the results of his experiments on spontaneous generation. Needham boiled mutton broth and then tightly stoppered the flasks. Eventually many of the flasks became cloudy and contained microorganisms. He thought organic matter contained a vital force that could confer the properties of life on nonliving matter. A few years later the Italian priest and naturalist Lazzaro Spallanzani (1729–1799) improved on Needham’s experimental design by first sealing glass flasks that contained water and seeds. If the sealed flasks were placed in boiling water for 3/4 of an hour, no growth took place as long as the flasks remained sealed. He proposed that air carried germs to the culture medium, but also commented that the external air might be required for growth of animals already in the medium. The supporters of spontaneous generation maintained that heating the air in sealed flasks destroyed its ability to support life. Several investigators attempted to counter such arguments. Theodore Schwann (1810–1882) allowed air to enter a flask containing a sterile nutrient solution after the air had passed through a red-hot tube. The flask remained sterile. Subsequently Georg Friedrich Schroder and Theodor von Dusch allowed air to enter a flask of heat-sterilized medium after it had passed through sterile cotton wool. No growth occurred in the medium even though the air had not been heated. Despite these experiments the French naturalist Felix Pouchet claimed in 1859 to have carried out experiments conclusively proving that microbial growth could occur without air contamination.
This claim provoked Louis Pasteur (1822–1895) to settle the matter once and for all. Pasteur first filtered air through cotton and found that objects resembling plant spores had been trapped. If a piece of the cotton was placed in sterile medium after air had been filtered through it, microbial growth appeared. Next he placed nutrient solutions in flasks, heated their necks in a flame, and drew them out into a variety of curves, while keeping the ends of the necks open to the atmosphere .Pasteur then boiled the solutions for a few minutes and allowed them to cool. No growth took place even though the contents of the flasks were exposed to the air. Pasteur pointed out that no growth occurred because dust and germs had been trapped on the walls of the curved necks. If the necks were broken, growth commenced immediately. Pasteur had not only resolved the controversy by 1861 but also had shown how to keep solutions sterile. The English physicist John Tyndall (1820–1893) dealt a final blow to spontaneous generation in 1877 by demonstrating that dust did indeed carry germs and that if dust was absent, broth remained sterile even if directly exposed to air. During the course of his studies, Tyndall provided evidence for the existence of exceptionally heat-resistant forms of bacteria. Working independently, the German botanist Ferdinand Cohn (1828–1898) discovered the existence of heat-resistant bacterial endospores
 The Spontaneous Generation Experiment. Pasteur’s swan neck flasks used in his experiments on the spontaneous generation of microorganisms.
The Spontaneous Generation Experiment. Pasteur’s swan neck flasks used in his experiments on the spontaneous generation of microorganisms.
2. Disprove of Spontaneous Generation theory
At that time, the age old idea of “Spontaneous Generation theory” was the dominant one. The idea that organism originate directly from non-living matter. (Life from non-living) also called as abiogenesis (a – not; bio – life; genesis – origin).
Ex : Maggots were developed spontaneously via recombination of matters in rotting materials. (ex meat)
The microbiology starts when the disprove of SG theory.
Louis Pasteur (1822 – 1895) and disproval of Spontaneous generation theory
He performed “gooseneck experiment”. The nutrient of flask was heated and the untreated – unfiltered air could pass in or out, but the germs settled in the gooseneck and no microbes were observed in the nutrient solution.
His concept of Germs theory of disease (means germs are responsible for the disease not the inert mater) ends the SG theory.
Contributions of Louis Pasteur (1822 – 1895)
- Disproved the SG theory
- Discovered that fermenting fruit to alcohol by microbes – From now the Fermentation started
- Sorted different microbes giving different taste of wine.
- He selected a particular strain (Yeast) for high quality wine.
- He developed a method to remove the undesired microbes from juice without affecting its quality. Heating the juice at 62.8°C for half-an hour did the job. This technique is called as Pasteurization, which is commonly used in the field of milk industry.
- He discovered that parasites (protozoa) causing pebrine disease of silk worm. He suggested that disease free caterpillars can eliminate the disease.
- He isolated the anthrax causing bacilli from the bloods of cattle, sheep and human being.
- He also demonstrated the virulence (ability of microbe to cause disease) of bacteria
- He developed vaccine (a killed or attenuated microbe to induce the immunity) against rabbis from the brains and spinal cord of rabbit
John Tyndall (1820 -1893)
- Proved that dust carries the germs and if no dust in the air, the sterile broth remained free of microbial growth for indefinite period.
- He also developed a sterilization method “Tyndallization”, referred as intermittent or fractional sterilization. The subsequent cooling and heating by steam for 3 days will remove the germs and their spores.
Martinus Willium Beijerinck (1851 – 1931)
- Developed the enrichment technique to isolate various group of bacteria.
- Isolated sulphur reducing bacteria and sulphur oxidizing bacteria from soil
- Isolated free-living nitrogen fixing bacterium, Azotobacter from soil,
- Root nodulating bacterium, Rhizobium, Lactobacillus, green algae were identified by him
- He confirmed the Tobacco mosaic virus causes disease and it incorporated in the host plant to reproduce.

Sergei Winogradsky (1856 – 1953)
The following are the contributions of Winogradsky to soil microbiology.
- Microorganisms involved in N cycle, C cycle, S cycle
- Nitrification process in soil
- Autotrophic nutrition of bacteria
- Chemolithotrophic nutrition of soil bacteria
- Discovered anaerobic nitrogen fixing bacterium Clostridium pasteurianum

Walther Hesse & Fannie E. Hesse (1883)
They used agar instead of gelatin for preparation of media. Agar goes to solution at 100°C and solidifies at 45°C. Till now this was not replaced by any other substance.
Joseph Lister (1878)
Developed Pure culture technique. Pure culture referred as the growth of mass of cells of same species in a vessel. He developed the pure cultures of bacteria using serial dilution technique.
He also discovered that carbolic acid to disinfect the surgical equipments and dressings leads the reduction of post-operational deaths/infections.
Alexander Fleming (1928) identified Penicillium notatum inhibiting Staphylococcus aureus and identified the antibiotic Penicillin
- 1929-Discovered antibiotic penicillin –important milestone in medical microbiology
- Found that natural substances having antimicrobial activity- Saliva,Nasal mucous
- Worked on Staphylococcus aureus,-inhibition of growth-due to Penicillin
- Florey &Chain-isolated Penicillin in pure culture.

Selman A Waksman, 1945 identified Streptomycin antibiotic from soil bacterium. He also coined the term antibiotics (referring a chemical substance of microbial origin which is in small quantity exert antimicrobial activity.
- 1927- Wrote the book on Principles of soil Microbiology
- In 1939 Waksman and his colleagues undertook a systematic effort to identify soil organisms producing soluble substances that might be useful in the control of infectious diseases, what are now known as antibiotics
- Within a decade ten antibiotics were isolated and characterized,
- three of them with important clinical applications
- actinomycin in 1940, streptomycin in 1944, and neomycin in 1949.
- Eighteen antibiotics were discovered under his general direction.

| Download this lecture as PDF here |
`
Introduction
Bacteria are mostly unicellular organisms that lack chlorophyll and are among the smallest living things on earth—only viruses are smaller. Multiplying rapidly under favorable conditions, bacteria can aggregate into colonies of millions or even billions of organisms within a space as small as a drop of water.The Dutch merchant and amateur scientist Anton van Leeuwenhoek was the first to observe bacteria and other microorganisms. Using single-lens microscopes of his own design, he described bacteria and other microorganisms (calling them “animacules”) in a series of letters to the Royal Society of London between 1674 and 1723.
Bacteria are classified as prokaryotes. Broadly, this taxonomic ranking reflects the fact that the genetic material of bacteria is contained in a single, circular chain of deoxyribonucleic acid (DNA) that is not enclosed within a nuclear membrane. The word prokaryote is derived from Greek meaning “prenucleus.” Moreover, the DNA of prokaryotes is not associated with the special chromosome proteins called histones, which are found in higher organisms. In addition, prokaryotic cells lack other membrane-bounded organelles, such as mitochondria. Prokaryotes belong to the kingdom Monera. Some scientists have proposed splitting this designation into the kingdoms Eubacteria and Archaebacteria. Eubacteria, or true bacteria, consist of more common species, while Archaebacteria (with the prefix archae—meaning ancient) represent strange bacteria that inhabit very hostile environments. Scientists believe these bacteria are most closely related to the bacteria which lived when the earth was very young. Examples of archaebacteria are those bacteria which currently live in extremely salty environments or extremely hot environments, like geothermal vents of the ocean floor
Microbes are organisms that we need a microscope to see. The lower limit of our eye’s resolution is about 0.1 to 0.2 mm or 100 – 200 um. Most microbes range in size from about 0.2 um to the 200 um upper limit, although some fruiting bodies of fungi can become much larger. Microbes include the bacteria, algae, fungi, and protozoa. In this lecture we will discuss mostly the bacteria and the fungi.
Bacteria are found everywhere in water, soil, and even air. These small prokaryotic cells, typically from 0.2 to 1 um in length, are capable of living in boiling water, frozen ground, acid volcanoes, and at the bottom of the ocean. They can reproduce by doubling with a generation time of 20 minutes, or survive for centuries in a resting stage. In natural waters (lakes, streams, oceans) their generation time is around 1 day. In soils they live in a film of water around plant roots or other particles, and their activity is dependent on the temperature and the amount of available moisture. In general, bacteria are found in concentrations of 106 cells/mL of water in surface waters, and 109 cells/mL of soil in soils and sediments.
Robert Koch (1843 -1910): The Father of Microbial Techniques
Robert Koch, a German Physician, is well known to the world of microbiology for this significant contributions especially in the area of microbial techniques. He introduced analine dyes for staining bacteria; used agar-agar and gelatin to prepare solid culture media; stressed the need for pure culture to study microbes in details; confirmed germ theory of disease, and laid down Koch’s postulates to test the pathogenesity of causative agents. He also discovered the casual organisms of anthrax disease of cattle (Bacillus anthracis) and tuberculosis (Mycobacterium tuberculosis).
Robert Koch was particularly concerned with this problem and, at first, he cultured bacteria on solid fruits and vegetables such as slices of boiled potato but many bacteria did not grow on such substrates. Then he perceived that it would be far better if a well-tried liquid medium could be solidified with some clear substance. Koch (1881) tried gelatin as a solidifying agent and succeeded in developing solid culture media, but gelatin, the first solidifying agent used, had serious disadvantage of becoming liquid above 28-30°C which is below the optimum temperature for the growth of human disease producing bacteria.
However, Koch replaced gelatin by agar in 1883-84 on the recommendation of F.E. Hesse, a German housewife, who had gained experience with the characteristics of agar in the process of making jelly. Agar is still frequently used as solidifying agent in microbiological laboratories. The development of solid culture media to grow pure culture was of fundamental importance and may be considered one of the Koch’s greatest contributions.
Besides developing solid culture media using gelatin and agar, Koch also evolved methods to placed microbes on glass slides and colour them with analine dyes (stains) so that the individual cells could be seen more clearly under the microscope.
KOCH’S POSTULATES
1. The microorganism must be present in every case of the disease but absent from healthy organisms.
2. The suspected microorganism must be isolated and grown in a pure culture.
3. The same disease must result when the isolated microorganism is inoculated into a healthy host.
4. The same microorganism must be isolated again from the diseased host.
“One microbe, one disease”
- Robert Koch (1843-1910) was the first to rigorously demonstrate that a specific disease was caused by a specific microorganism.
- Koch worked on anthrax, a disease mainly of animals. Koch noticed that cattle that died of anthrax all seemed to have a certain rod-shaped bacterium in blood, not found in healthy animals. Koch was able to isolate the bacterium in pure culture, put it back into healthy cows, and reproduce the disease.
- Koch’s Postulates: a logical way to identify the microbe causing a disease
- A specific microbe must be present in all disease cases
- Microbe must be cultivated outside host in a pure culture
- When pure culture of microbe is inoculated into healthy hosts, disease symptoms identical to those of initial host must be reproduced
- Microbe can be isolated again in pure culture from this experimentally inoculated host.
- Initial attempts to isolate microbes used sliced potatoes or nutrient media containing gelatin — not ideal media. Then Fannie Hesse (wife of lab worker) suggested agar, a gelling agent used in cooking. Agar rapidly became the standard gelling agent for microbial isolation because it is relatively inert (only some marine microbes have enzymes to digest agar). Agar only melts at high temperatures (100oC); once melted, it remains liquid until about 45oC, at which point it gels.
- Koch’s success at identifying anthrax with bacterium Bacillus anthracis led both Koch and Pasteur to identify the causes of many diseases — cholera, tuberculosis, plague, etc. — over the next few decades (late 1880’s) — the “Golden Age of Microbiology” (~ 1870-1920). Note that many microbiologists would regard the present as a new “Golden Age”, since the development of molecular biological techniques, PCR, molecular phylogeny, and other developments have revealed many new insights and opened a world of new research directions and ways of understanding microbes.
| Download this lecture as PDF here |
The control of microbial growth is necessary in many practical situations, and significant advances in agriculture, medicine, and food science have been made through study of this area of microbiology. The microorganisms are ubiquitous in nature. In order to study the nature and characteristics of a particular microbe, it is essential to isolate it from other contaminating microorganisms. This can be achieved by maintaining a completely sterile environment in which the microbe of interest is selectively grown. It is necessary that not only the place you are working with microorganisms should be free from contamination (other living organisms) but, the media and the materials you are using to handle and grow specific microorganisms should be free from other microbial contaminants. For this purpose ‘sterilization’ of the place of work materials and media have to be done.
“Control of growth” as used here means to prevent the growth of microorganisms. This control is affected in two basic ways: (1) by killing microorganisms or (2) by inhibiting the growth of microorganisms. Control of growth usually involves the use of physical or chemical agents which either kill or prevent the growth of microorganisms. Agents which kill cells are called cidal agents; agents which inhibit the growth of cells (without killing them) are referred to as static agents. Thus the term bactericidal refers to killing bacteria and bacteriostatic refers to inhibiting the growth of bacterial cells. A bactericide kills bacteria; a fungicide kills fungi, and so on.
Sterilization is a process of complete removal or killing of all forms of microbial life including spores from an object, surface, medium or environment without spoiling its nature.
Methods
There are various sterilization techniques available. However, several factorsinfluence the effectiveness of sterilization process like, the concentration of antimicrobial agents, time and temperature of exposure, size of population, type of contaminating microbes etc.
Sterilization is brought about by a combination of physical and chemical agents that adversely affect the microorganisms either by causing damage to the cell wall or cell membrane or by inactivating the enzymes or by interfering with the synthesis of nucleic acids and protein.
I. PHYSICAL AGENTS
There are different types of physical agents.
(i) Heat: The heat employed for removal of micro-organisms varied with the nature of object and also depend on the purpose. Based on these different processes are employed.
(a) Moist heat
It is the widely used effective means of sterilization process. In this, steam under high pressure is employed which imparts high penetration power resulting in the hydration of cells and coagulation of protein leading to the death of the microorganism. Autoclave is the apparatus used for sterilization by moist heat. The autoclave is a double-jacketed steam chamber. The chamber is equipped with a device for generating saturated steam. It can be maintained at a particular temperature and pressure for any period of time. During operation of autoclave the air in the chamber is evacuated by steam since presence of air will reduce the temperature in the chamber. The time required for sterilization will depend upon the materials to be sterilized. Solid materials must be heated for a longer time (1-2 hours) while liquid media can be sterilized within 15-30 minutes. Also acidic materials require shorter period than alkali materials. A temperature of 121°C for 15 min at a pressure of 15 lbs/ sq.inch is the sterilizing condition in the autoclave.
The autoclave is a double-jacketed steam chamber. The chamber is equipped with a device for generating saturated steam. It can be maintained at a particular temperature and pressure for any period of time. During operation of autoclave the air in the chamber is evacuated by steam since presence of air will reduce the temperature in the chamber. The time required for sterilization will depend upon the materials to be sterilized. Solid materials must be heated for a longer time (1-2 hours) while liquid media can be sterilized within 15-30 minutes. Also acidic materials require shorter period than alkali materials. A temperature of 121°C for 15 min at a pressure of 15 lbs/ sq.inch is the sterilizing condition in the autoclave.
Advantages
Steam can penetrate through materials and sterilization is achieved by the coagulation or denaturation of proteins and other cell constituents. Liquid media, solid media, laboratory equipments (cloth, glasswares, etc.,) can be sterilized. The temperature and pressure is high enough to kill spores, vegetative cells and viruses.
Disadvantages
Temperature sensitive media, animal tissue culture media, antibiotics, amino acids, cannot be sterilized. Sometimes water may get inside incase of improper packing.
(b) Dry heat
This process is accomplished in a hot-air oven. Hot air or dry heat is employed for sterilization. The dry heat penetrates substances more slowly than the moist heat. Hence, the time required for effective sterilization is long (2 to 3 hours) and also the temperature required is too high (160°C -180°C). Microbial death results from the oxidation of cell constituents.
Advantages
Dry heat does not corrode glassware and metal instruments as moist heat does. All glassware’s can be sterilized.
Disadvantages
The sterilization process is slow. It is not suitable for heat sensitive materials like many plastic and rubber items.
(c) Boiling at 100°C for 30 minutes. Kills everything except some endospores (Actually, for the purposes of purifying drinking water 100o for five minutes is probably adequate though there have been some reports that Giardia cysts can survive this process). To kill endospores, and therefore sterilize the solution, very long or intermittent boiling is required.
(d) Pasteurization is the use of mild heat to reduce the number of microorganisms in a product or food. In the case of pasteurization of milk the time and temperature depend on killing potential pathogens that are transmitted in milk, i.e., staphylococci, streptococci, Brucella abortus and Mycobacterium tuberculosis. For pasteurization of milk:
batch method (Low temperature holding): 62.8oC for 30 minutes flash method (High temperature short time): 71.7oC for 15 seconds
(e) Intermittent sterilization or Tyndallization is the process of boiling the materials at 100 oC for 30 min. successively for three consecutive days. Destroys vegetative cells and spores; germinated spores.
(f) Incineration burns organisms and physically destroys them. Incineration is the complete burning of the material in to ashes. Used for needles, inoculating wires, glassware, etc. and objects not destroyed in the incineration process. This is the direct and ultimate method of destroying cells. It is achieved by keeping the materials directly in contact with the flame of Bunsen burner as a result all the microorganisms in the surface are destroyed completely. Inoculating loops, needles and spreading rods are sterilized by this method.
Advantages: Immediate and quick.
Disadvantages: Cannot be used to sterilize heat labile material, material is lost by incineration.
Recommended use of heat to control bacterial growth
Treatment | Temperature | Effectiveness |
Incineration | 5000 C | Vaporizes organic material on non flammable Surfaces but may destroy many substances in the process |
Boiling | 100 0C | 30 minutes of boiling kills microbial pathogens and vegetative forms of bacteria but may not kill bacterial endospores |
Intermittent boiling | 1000C | Three 30-minute intervals of boiling, followed by Periods of cooling kills bacterial endospores. |
Autoclave and pressure cooker (steam under pressure) | 1210C for 15 min. | Kills all forms of life including bacterial endospores. The substance being sterilized must be maintained at the effective T for the full time |
Dry heat (hot air oven) | 1600 C /2 hours | For materials that must remain dry and which are not destroyed at the between 121oC and 170oC Good for glassware, metal, not plastic or rubber items |
Dry heat (hot air oven) | 1800 C /1 hour | Same as above. Note increasing T by 10 degrees shortens the sterilizing time by 50 percent |
Pasteurization | 62.8 0C /30 min. | kills most vegetative bacterial cells including pathogens such as streptococci, staphylococci and Mycobacterium tuberculosis |
Pasteurization | 71.7 0C/15 seconds | Effect on bacterial cells similar to batch method; for milk, this method is more conducive to industry and has fewer undesirable effects on quality or taste |
(ii) Radiation
Energy transmitted through space in a variety of forms is generally called radiation. It is also known as “cold sterilization” as only little heat is produced during the process. The most significant of this is electromagnetic radiation. The energy content and radiation wavelength are inversely proportional to each other. Radiation may be ionizing or non-ionizing.
Ionizing radiation
High-energy electron beams (Gamma, X-rays, alpha and beta particles) have sufficient energy to cause ionization of molecules. They drive away electrons and split the molecules into ions. Water molecules are split into hydroxyl radicals (OH-), electrons and hydrogen ions (H+). OH- ions are highly reactive and destructive to normal cellular compounds such as DNA and proteins. Thus ionizing radiations are used in sterilization.
e.g. 36Cs, 60Co
Advantages: X-rays and Gamma rays have high penetrating power. Packed food and medical equipments are sterilized by using x-rays and gamma rays.
Disadvantage: Generating and controlling X-rays for sterilization is highly expensive.
Non-ionizing radiation
This includes ultraviolet (UV) rays. UV at a wavelength of 265 nm is most bactericidal. Absorption of UV radiation produces chemical modification of nucleoproteins i.e., thymine dimer formation that leads to misleading of genetic codes. This mutation impairs the total functions of the organism, consequently causing its death.
Advantages
It is used to maintain aseptic conditions in laminar air flow chamber, lab, hospitals, pharmaceuticals, industries etc., and also in the sterilization of water and air.
Disadvantage:
UV radiation has very little ability to penetrate matter and hence the micro organisms on the surface of an object are destroyed.
III) Filtration
Filtration involves the passage of liquid or gas through a screen like material that has spores small enough to retain the micro organism of certain size. It is used to sterilize heat sensitive substance like enzyme solutions, bacterial toxins, certain biological media, cell extract and some sugars. Various types of filters are available in different grades of porosity. Vacuum or pressure is required to move the solutes through the filter.
Involves the physical removal of all cells in aliquid or gas, especially important to sterilize solutions which would be denatured by heat (eg: antibiotics, injectable drugs, amino acids, vitamins etc.)
Advantages:
It si the best way to reduce microbial population in solutions of heat sensitive materials and it is sued to sterilize liquid media, vitamin solutions, hormones, growth factors, enzymes.
Disadvantages
Pleomorphic structures like mycoplasma cannot be effectively filtered by this technique. It is applicable to sterilize only small quantities.
Commonly used filters in micro biology
The sintered glass fliter is made of fused Jen or pyrex glass, manufactured in such a way as to be porous, with apore size and adsorptive charge sufficient to retain bacteria. The seitz filters are compressed asbestos discs having porosity sufficiently small to retain bacteria. Tye chamber land filters are made of porcelain. The mandler/berkfield filters are made of diatomaceous earth. The membrane filter is a cellulose or nitrocellulose membrane with apore size sufficiently small (0.01mm to 10 mm) to trap and thereby remove bacterial from a liquid. The membrane filters are also used to concentrate and trap the micro organisms in water and other liquids. HEPA (High efficiency particulate air filters are of fibre galss filters for sterilization of air.
Low temperature
Most organisms grow very little or not at all at 0O C. Store perishable foods at low temperatures to slow rate of growth and consequent spoilage (eg: milk). Low temperatures are not bactericidal. Psychrotrophs, rather tah true psychrophiles are the usual cause of food spoilage in refrigerated foods.
Dessication / Drying (removal of H2O)
Most micro organisms cannot grow at reduced water activity (aw < 0.90). Often used to preserve foods (eg: fruits, grains etc). methods involve removal of water from product by heat, evaporation, freeze drying, addition of salt or sugar.
Surface tension
is a property of the surface of a liquid that allows it to resist an external force. It is revealed, for example, in floating of some objects on the surface of water, even though they are denser than water, and in the ability of some insects (e.g. water striders) and even reptiles (basilisk) to run on the water surface. This property is caused by cohesion of like molecules, and is responsible for many of the behaviors of liquids.
Surface tension has the dimension of force per unit length, or of energy per unit area. The two are equivalent—but when referring to energy per unit of area, people use the term surface energy—which is a more general term in the sense that it applies also to solids and not just liquids.
In materials science, surface tension is used for either surface stress or surface free energy
Osmotic pressure – plasmolysis/ plasmotysis
Is the process in plant cells where the plasma membrane pulls away from the cell wall due to the loss of water through osmosis. The reverse process, cytolysis, can occur if the cell is in a hypotonic solution resulting in a higher external osmotic pressure and a net flow of water into the cell. Through observation of plasmolysis and deplasmolysis it is possible to determine the tonicity of the cell’s environment as well as the rate solute molecules cross the cellular membrane.
Chemical agents
Chemical that is used to kill or inhibit the growth and development of micro organisms are called anti microbial agents. Disinfectants and antiseptics come under anti microbial agents and are usually used on inanimate materials. The mechanism of action is complex and non specific. It may act on lipid portion of cell membrane, oxidize or reduce an important functional group of an enzyme, prevent certain bio synthesis or cause extensive breakdown of DNA.
Types of microbial agents
Chemical sterilants
Chemical sterilants are chemical anti microbial agents that are usd fro sterilization of heat sensitive substance/ materials. Normally plastic petriplates and medical supplies such as blood transfusion sets, plastic syringes, lenses etc. could be sterilized even in packets or bundles using ethylene oxide, formaldehyde or formalin is effectively used to sterilize enclosed areas/a septic chambers at 22 O C with a relative humidity of 60 – 80 %.
Antispetics
Microbicidal agents harmless enough to be applied to the skin and mucous membrane, should not be taken internally. Eg: mercurails, silver nitrate, iodine solution, alcohols, detergents.
Disinfectants
Agents that kill micro organisms, but not necessary their spores, not safe for application to living tissues, they are used on inanimate objects such as tables, floors, utensils etc. eg: Chlorine, hypochlorites, chlorine compounds, Lysol, copper sulfate, quaternary ammonium compounds.
Phenol
Derivative of phenol like benzyl resorcinol, o-cresol, m-cresol, etc., are used as effective disinfectants 5% aqueous solutions of phenols are used as disinfectant. It alters the protein structure and leads to denaturation of proteins and enzymes. Also affects permeability of cytoplasmic membrane. They readily kill vegetative cells of bacteria and fungi but for spores.
Alcohol
Alcohol at 70% concentration is more effective. It brings about denaturation and coagulation of protein. Ethanol is routinely used in laboratories to surface sterilize worktables and hands of the researcher/ experiment.
Halogens
Halogens such as hypocholrites, choramines and povidone- iodine are used to sanitize utensils, surface sterilize in animate objects, table surfaces and other instruments.
Heavy metals
Heavy metals such as mercuric chloride are also used for surface sterilization purposes. Heavy metals acts as oxidizing agents and kill the micro organisms on the surface of the object. Usually 0.1 % mercuric chloride is used in the laboratories to sterilize the surface of worktable and explants.
Detergents
Detergents are those compounds that make water repellant surfaces more wettable. There are two types of detergents viz., ionic and non ionic. Detergent soaps and other synthetic detergents are used for washing/cleaning glass wares, table tops etc.,
Common antiseptics and disinfectants
| Chemical | Action | Uses |
Ethanol (50 -70 %) | Denatures proteins and solubilizes lipids. | Anti septic used on skin |
Isopropanol (50 – 70 %) | Denatures proteins and solubilizes lipids. | Anti septic used on skin. |
Formaldehyde (8%) | Reacts with NH2, SH and COOH groups. | Disinfectant, kills endopsores. |
Tincture of Iodine ( 2% in 70 % alcohol) | Inactivates proteins | Antiseptic used on skin |
Chlorine (Cl2) gas | Forms hypochlorous acid (HClO), a strong oxidizing agent. | Dis infect drinking water, general disinfectant. |
Silver Nitrate (Ag No3) | Precipitates proteins. | General antiseptic and used in the eyes of newborns. |
Mercuric chloride | Inactivates proteins by reacting with sulfide groups. | Disinfectant although occasionally used as an antiseptic on skin. |
Detergents (eg: Quaternary ammonium compounds) | Disrupts cell membranes. | Skin antiseptics and disinfectants. |
Chemotherapeutic agents
Antimicrobial agents of synthetic origin useful in the treatment of microbial or viral disease. Examples: sulfonilamides, isoniazid, ethambutol, AZT, chloramphenicol.
Antibiotics
Antimicrobial agents produced by micro organisms that kill or inhibit other micro organisms. This is the microbiologist’s definition. A more broadened definition of an antibiotic includes many chemical of natural origin which has the effect to kill pr inhibit the growth of other types cells. Since most clinically useful anti biotics are produced by micro organisms and are used to kill or inhibit infectious bacteria we follow classic definition.
Antibiotics are low molecular weight (non- protein) molecules produced as secondary metabolites, mainly by micro organisms that live in the soil. Most of these micro organisms form some type of a spore or other dormant cell, and there is thought to be some relationship between anti biotic production and the process of sporulation. Among the molds, the notable antibiotic producers are penicillium and cephalosporium, which are the main source of the beta lactam antibiotics. In the bacteria, the actinomycetes, notable streptomyces species, produce a variety of types of anti biotics including the aminoglycosides (eg: streptomycin), macrolides (eg: erythromycin) and the tetracycline. Endospore forming bacillus species produce polypeptide anti biotics such as polymyxin and bactracin.
Chemical class | examples | Biological source | Spectrum (effective against) | Mode of action |
Beta – lactams | Penicillin G, Cephalothin | Penicillium notatum and cephalosporium sp. | Gram positive bacteria | Inhibits steps inc ell wall (peptidoglycan) synthesis and murein assembly. |
Aminoglycosides | streptomycin | Streptomyces griseus | Gram positive and gram negative bacteria | Inhibit translation (protein synthesis) |
glycopeptides | vancomycin | Streptomyces orientales | Gram positive bacteria, esp. staphylococcus aurues | Inhibits steps inn murein (peptidoglycan) biosynthesis and assembly |
macrolides | erythromycin | Streptomyces erythreus | Gram positive and gram negative bacteria not enteric,. Neisseria, legionella, mycoplasma | Inhibits translation(protein synthesis) |
polypeptides | polymyxin | Bacillus polymyxa | Gram negative bacteria | Damages cytoplasmic membranes |
Polyenes | amphotericin | Streptomyces nodosus | Fungi | Inactivate membranes containing sterols |
tetracyclines | tetracycline | Streptomyces sp | Gram positive and gram negative bacteria, rickettsias | Inhibit translation (protein synthesis) |
Chloramphenicol | Chloramphenicol | Streptomyces venezuelae | Gram positive and gram negative bacteria | Inhibit translation (protein synthesis) |
| Download this lecture as PDF here |
Microbial Metabolism
Metabolism refers the sum of biochemical reactions required for energy generation and the use of energy to synthesize cellular materials.
The energy generation component is referred as catabolism and the build up of macromolecules and cell organelles are referred as anabolism.
During catabolism, the energy is changed from one compound to
another and finally conserved as high energy bonds of ATP.
ATP is the universal currency for energy. When energy is required for anabolism, it may be sent as high energy bonds of ATP which has the value of 8 kcal per mole.
Based on the source of carbon, the microbes can be divided into two groups namely, autotrophs and heterotrophs. Autotrophs utilize CO2 as sole carbon source and heterotrophs use organic carbon as sole carbon source.
I. Energy generation by heterotrophs
Heterotrophs use variety of carbon sources. Glucose is being the simple and wide variety of microbes prefers it. The glucose can be taken up by bacterium through diffusion and can be readily utilized. There are three possible pathways available in bacteria to use glucose. All these path ways are fermentative type and substrate level phosphorylation occurs.
- Embden-Meyerhof path way
- Phosphoketolase path way
- Entner – Doudoroff path way.
SUBSTRATE LEVEL PHOSPHORYLATION: (Fermentation)
The EMP pathway, phosphoketolase pathway and ED pathway end with one or two ATP synthesis by substrate level phosphorylation. There won’t be any external source of electron acceptor will come in these reactions.  A. Embden-Meyerhof path way
A. Embden-Meyerhof path way
This is the path way of glycolysis most familiar and common to most of the organisms. The path way is operated by yeast to produce alcohol and lactic acid bacteria to produce lactic acid and several organic acids, gases, fatty acids, and alcohols. The path way is as follows:
Glucose à 2 pyruvate + 2 ATP + 2 NADH2
After pyruvate is formed, if the organism is a respirative type, the pyruvate will go to Krebs cycle and if the organism is fermentative, the reduction process ends with organic acids, alcohols etc.
A model fermentation:After an intermediate product, a reduction takes place in fermentation, whereas, if respiration, CO2 will be formed by complete oxidation through Krebs’s cycle
(Note: After pyruvate, the reduction process leads to fermentation and complete oxidation leads to respiration)
The Embden – Meyerhof path way can lead to a wide array of end products depending on the path ways taken in the reductive steps after the pyruvate formation.
The following are some of the such fermentations:
| Fermentation | End products | Model organism |
| Homolactic fermentation | Lactic acid | Lactobacillus |
| Mixed acid fermentation | Lactate, acetate, formate, succinate | Enterobater |
| Butyric acid fermentation | Butric acid, acetone | Clostridium acetobutylicum |
| Propionic acid fermentation | Propionic acid | Propionibacterium |
| Alcohol fermentation | Ethanol | Saccharomyces |
B. Phosphoketolase path way (Heterolactic path way)
The phosphoketolase path way is distinguished by the key cleavage enzyme phosphoketolase, which cleaves pentose to glyceroldehyde 3 phosphate and acetyl phosphate. The path way ends with ethanol and lactic acid. Ex. Lactobacillus, Leuconostoc. The overall reaction is,
Glucose à 1 lactate + 1 ethanol + 1 CO2 + 1 ATP
This path way is useful in the dairy industry for preparation of kefir (fermented milk), yogurt, etc.
C. Entner – Doudoroff pathway
Only few bacteria like, Zymomonas mobilis employ the ED pathway. The path way is as follows:
The overall reaction is
Glucose à 2 ethanol + 2 CO2 + 1 ATP
The alcohol productivity of Zymomonas is higher than yeast because of this fermentative pathway.
( Note : All the three pathways are end with 1 or 2 ATP by substrate level phosphorylation by means fermentation)
OXIDATIVE PHOSPHORYLATION (Respiration)
If the organism is a respiratory type (that means complete oxidation of glucose), it needs four essential metabolic components for their respiration and oxidative phosphorylation.
a. Tricarboxylic acid cycle (also known as citric acid cycle or Kreb’s cycle) The pyruvate formed during glycolysis will be completely oxidized to 3 CO2 by the use of this cycle. During oxidation of one pyruvate through TCA cycle, 4 NADH2, 1 FADH2 and 1 GTP are produced along with 3 CO2.
b. A membrane and associated Electron Transport System (ETC) The electron transport chain is a sequence of electron carriers transport the electrons to a terminal electron acceptor. During this flow of electron in the membrane, a proton motive force across the membrane leads to formation ATP (is referred as electron transport phosphorylation).
c. An outside electron carrier: for aerobic respiration, O2 is the terminal electron acceptor and reduced to H2O. This is normal for higher organisms. But in anaerobic bacteria, the terminal electron acceptor may be of nitrite, nitrate, sulphate or carbon dioxide.
d. A membrane bound ATPase enzyme: The proton motive force developed during ETC leads to formation of ATP by enzyme ATPase present in the membrane. (As in the diagram)



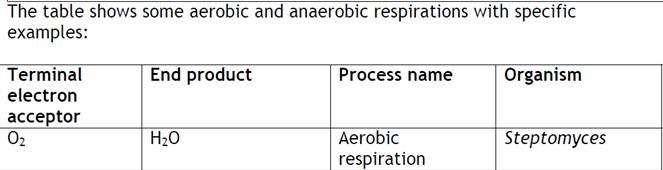
The table shows some aerobic and anaerobic respirations with specific examples:
| Terminal electron acceptor | End product | Process name | Organism |
| O2 | H2O | Aerobic respiration | Streptomyces |
| NO3 | NO2, N2 | Denitrification | Pseudomonas denitrificans |
| SO4 | S or H2S | Sulphate reduction | Desulfovibrio desulfuricans |
| Fumarate | Succinate | Anaerobic respiration | Escherichia |
| CO2 | Methane (CH4) | Methanogenesis | Methanococcus |
In aerobic organisms, the terminal electron acceptor will be of O2. In some anaerobic organisms, after the electron transport chain, instead of O2, some inorganic compounds like sulphate, nitrate or some organic compounds like fumarate act as terminal electron acceptor. Such type of respiration is referred as anaerobic respiration and the normal O2 mediated respiration is referred as aerobic respiration. The above table shows some anaerobic respiration with some terminal electron acceptors. The process is named based on the compounds as sulphur reduction, denitrification and methanogenesis.
Energy generation by autotrophs
Autotrophs use CO2 as their sole carbon source. There are two types such as photoautotrophs and chemoautotrophs. Photoautotrophs use light as energy source and CO2 as carbon source. Chemoautotrops use chemicals (especially inorganic) as energy source and CO2 as carbon source.
I. Energy and carbon assimilation by photoautotrphs: (Photoautotrophy)
Phototrophs use sunlight to produce ATP through phosphorylation, referred as photophosphorylation. The phototrophs convert the light energy to chemical energy (ATP) through the process called photosynthesis.
Photosynthesis is a type of metabolism in which catabolism and anabolism occur as sequence. The catabolic reaction (energy generating process) of photosynthesis is light reaction in which the light energy is converted to chemical energy (ATP) and electrons or reducing powers (NADPH). The anabolic reaction (macromolecule synthesis) of photosynthesis is dark reaction in which CO2 is converted to organic molecules (carbohydrates), which is also called as CO2 fixation.
For conversion of light energy to ATP, the bacteria possess light harvesting pigments. They are chlorophyll a, carotenoids, phycobiliproteins (which are present in cyanobacteria) and bacteriochlorophyll (which are present in purple sulphur bacteria). In bacteria, there are two types of light reactions (conversion of light to ATP) and two types of CO2 fixation occur.
A. Light reaction (Photophosphorylation)
For photophosphorylation, light harvesting pigments, a membrane electron transport chain, source of electron (electron donor) and ATPase enzymes are required. Two types of photophosphorylations occur during photosynthesis. They are cyclic photophosphorylation and non-cyclic photophosphorylation.
- In plant and cyanobacteria, both cyclic and non-cyclic photophosphorylation occurs whereas in purple bacteria, the cyclic photophosphorylation only occurs.
- In plant and cyanobacteria, the electron source is water, by photolysis, H2O split into H+ and O2 and during the process, O2 is evolved and referred as oxygenic photosynthesis
- Since, the sulphur bacteria is an anaerobic bacterium, they use H2S instead of H2O as electron donor. Since, there won’t be any O2 evolution during photosynthesis, referred as anoxygenic photosynthesis.
Difference between plant and bacterial photosynthesis
| organisms | Plant photosynthesis | Bacterial photosynthesis |
| plants, algae, cyanobacteria | purple and green bacteria | |
| type of chlorophyll | chlorophyll a absorbs 650-750nm | bacteriochlorophyll absorbs 800-1000nm |
| Photosystem I (cyclic photophosphorylation) | present | present |
| Photosystem II (noncyclic photophosphorylation) | present | absent |
| Produces O2 | Yes (Oxygenic) | No (Anoxygenic) |
| Photosynthetic electron donor | H2O | H2S, other sulfur compounds or certain organic compounds |
1. The oxygenic photophosphorylation
The end product of the light reaction is ATP, NADPH and O2. The ATP and NADPH, the energy and electron sources thus produced were used for dark reaction.
2. The anoxygenic photo phosphorylation
The anoxygenic photo phosphorylation will take palce as in the image and the end product of the light reaction is ATP, NADPH and Sulphur. The ATP and NADPH, the energy and electron sources thus produced were used for dark reaction.
B. Dark reaction (CO2 fixation)
The dark reaction in which the ATP and NADPH were used as energy and electron sources to fix the CO2 as carbohydrates. The pathway involved in the dark reaction is Calvin cycle, by which the CO2 is fixed as phosphoglyceic acid and lead to formation of many sugars. The enzyme RuBiSCO is the key enzyme for this process.
The following pathway shows the Calvin cycle and the formation of key monomers for anabolic reactions such as hexose phosphate – polysaccharides; pyruvic acid – amino acid and fatty acid; pentose phosphate – DNA and RNA.


A complete model of light and dark reaction of photosynthesis
Another way of CO2 fixation by phototrophs
In phototrophs, the electron and energy were derived form sunlight and carbon from CO2 fixation through Calvin cycle. But some bacteria may derive electron and energy from sunlight and fixes CO2 by some other path way, not the Calvin cycle. The example is Photosynthetic green bacteria (Chlorobium). They derive NADPH and ATP through cyclic phosphorylation, but CO2 fixation is by reverse TCA cycle. Since TCA cycle is amphibolic pathway (referring the cycle can operate in both the directions), it can also be used to fix the carbon-di-oxide if operated reversely. The pathway is as follows:
Another way of CO2 fixation is by methanogens: They use CO2 as terminal electron acceptor and forms CH4 (methane). They also fix by acetyl CoA pathway for fixing CO2.
Synopsis:
| Organism | Light reaction/ATP generation | Dark reaction/CO2 fixation |
| Cyanobacteria (Nostoc), plant and alga | Cyclic and non-cyclic photophosphorylation Oxygenic photosynthesis | Calvin cycle |
| Purple bacteria (Chromatium) | Cyclic and non-cyclic photophosphorylation Oxygenic photosynthesis | Calvin cycle |
| Green bacteria (Chlorobium) | Cyclic and non-cyclic photophosphorylation Oxygenic photosynthesis | Reverse TCA cycle |
II. Energy and carbon assimilation by Chemoautotrophs: (Chemoautotrophy)
Since the chemoautotrophs use inorganic chemicals for their energy and electron source, they are referred as chemolithotrophs or chemolithotrophic autotrophs.
These organisms remove electron from an inorganic substance and put them through electron transport chain for ATP synthesis (through electron transport phosphorylation). At the same time, the electrons were also flow through reverse electron transport chain and with the end product of NADPH. These ATP and NADPH were used for CO2 fixation through Calvin cycle. These bacteria are obligate aerobic organisms. Some examples of the chemolithotrophs are as follows:
Groups of chemolithotrophs
| Physiological group | Energy source | Oxidized end product | Organism |
| hydrogen bacteria | H2 | H2O | Alcaligenes, Pseudomonas |
| nitrifying bacteria | NH3 | NO2 | Nitrosomonas |
| nitrifying bacteria | NO2 | NO3 | Nitrobacter |
| sulfur oxidizing bacteria | H2S or S | SO4 | Thiobacillus, Sulfolobus |
| iron oxidizing bacteria | Fe 2+ | Fe3+ | Gallionella, Thiobacillus |
The following diagram showing energy generation and CO2 fixation by different chemolithotrophs:
| Download this lecture as PDF here |
The energy captured within ATP can then be harnessed to create order in the form of biosynthetic reactions. In a hypothetical enzyme reaction that converts substrates A−H and B−OH to A−B and H2 O, the energy from ATP hydrolysis is first used to convert B−OH to a higher-energy intermediate, B−O−PO4. This compound is only transiently formed, with the energy released during its decay used by the enzyme to form A−B. Thus, the energy released from the ATP hydrolysis reaction (large −_G) is coupled to the synthesis reaction (large +_G). In this way, the cell can progressively create order.
Electron transport system (ETS)
- Although cells could transfer electrons directly from NADH to oxygen, this would liberate all energy in NADH directly as heat.
- NADH possesses lots of energy. If electrons are transferred directly to oxygen:
- NADH + O2 NAD + H2O, delta Go’ = – 218 kjoules/mole
- If NADH has ~218 kjoules of energy, and it only takes 30.5 kjoules to make one ATP, could conceivably make 218/30.5 = ~ 7 ATP per NADH if energy conversion were 100% efficient.
- In practice, cells have evolved ways to get up to 40% efficiency (~ 3 ATP/NADH) under optimal circumstances.
- Electron transport system (ETS) = membrane-bound pathway transferring electrons from organic molecules to oxygen.
- ETS moves both electrons and protons:electrons are passed from carrier to carrier in the membrane, while protons are moved from inside to outside of membrane
- Net result: electrons enter ETS from carriers like NADH or FADH, wind up at terminal oxidase, get attached to oxygen.
- ETS consists of 4 complexes, connected by mobile carriers (Coenzyme Q, cytochrome c) that shuttle between complexes in membrane

Specific carriers of ETS:
- mitochondria (in eukaryotes): NADH —> (Flavoprotein Iron sulfur proteins Quinone cytochrome b cytochrome c cytochrome a cytochrome a3 oxygen
- bacteria (prokaryotes) have different ETS carriers, shorter chains. In E. coli, can have two different terminal oxidases, one functions at high oxygen levels, one at lower oxygen levels. Cytochromes involved include: b558, b595, b562, d, and o
- proton gradient and oxidative phosphorylation (oxphos)
Chemiosmotic hypothesis (Peter Mitchell, 1961)
- As electrons flow through ETS, at certain steps protons (H+) are moved from inside to outside of the membrane.
- This builds up proton gradient; since + charges are removed from inside of cell, – charge remains inside, mainly as OH- ions.
- pH just outside membrane can reach 5.5, pH just inside membrane can reach 8.5 —> difference of 3 pH units, or 1000x concentration differential of H+ across membrane. This represents potential energy stored up in proton gradient = proton motive force.
- Membrane is basically impermeable to protons, so gradient doesn’t get squandered away by leaky reentry.
- ATP synthase protein complex contains only channels for proton entry. As protons push in through channel, the base rotates. Specific binding sites allow ADP + Pi ATP. This can be called chemiosmotic phosphorylation (assuming chemiosmotic hypothesis is correct), or oxidative phosphorylation (makes no assumption about mechanism.
Oxidative phosphorylation
Differences between respiration in mitochondria (eukaryotes) and bacteria (procaryotes)
- In Eukaryotes:
- ETS located in inner mitochondrial membrane. Proton gradient develops across inner mitochondrial membrane.
- Mitochondria are very efficient at generating proton gradient. Can measure how many ~P bonds (in ATP) are made for each O2 consumed = P/O ratio.
- With NADH as electron donor, P/O ratio can be 3 (means 3 ATP made per NADH).
- But with FADH as electron donor, P/O ration only 2 (fewer protons are transported, less proton gradient).
- Overall efficiency of respiration in mitochondria: ~ 40% (means that about 40% of energy in glucose actually gets converted to ATP).
- In Prokaryotes:
- ETS located in cytoplasmic membrane. Proton gradient develops across this membrane.
- Bacteria are not as efficient. ETS chains are shorter, P/O ratios are lower.
- As a ballpark estimate, P/O ratios for NADH are only ~2. Overall efficiency of glucose oxidation is closer to 28%, not 40%.
Inhibitors of Oxidative Phosphorylation
- Several chemicals can block electron transfer in ETS, or transfer of electrons to oxygen. All are strong poisons. Some examples:
- Carbon monoxide — combines directly with terminal cytochrome oxidase, blocks oxygen attachment
- Cyanide (CN-) and Azide (N3-) bind to cytochrome iron atoms, prevent electron transfer.
- Antimycin A (an antibiotic) inhibits electron transfer between cyt b and c.
Anaerobic respiration
- Use of acceptors other than oxygen.
- Most common in bacteria. Most alternative electron acceptors are inorganic molecules, but some organic molecules can serve.
- As with aerobic respiration, anaerobic respiration uses ETS, membrane localization, proton gradient, and ATP synthase.
- Processes are of great importance both ecologically and industrially.
Anaerobic respiration
Nitrate (NO3-).
- Process called denitrification. Also called dissimilative nitrate reduction. Reduced waste products are excreted in significant amounts.
- Redox potential is + 0.42 v (compared to + 0.82 v for oxygen). So organisms respiring anaerobically gain less energy than with oxygen.
- Requires new terminal oxidase called nitrate reductase. Enzyme is repressed by oxygen, synthesis turned on in absence of oxygen.
- Process can have several steps, proceed in two different directions:
- (A) nitrate (NO3-) nitrite (NO2-) ammonia (NH3)
- (B) nitrate (NO3-) nitrite (NO2-) nitrous oxide (N2O) dinitrogen gas (N2)
- Second process is major pathway for loss of nitrogen compounds from soil, return of nitrogen to atmosphere.
- Pseudomonas species are common denitrifiers, widespread in soils. When fertilized soils become flooded, oxygen is rapidly depleted, pseudomonads switch to anaerobic respiration and can use up soil nitrate, leaving field in unfertile state.
- Note: Studied this in lab. Media must contain nitrate in addition to nutrients, otherwise won’t work. Also, in scavenger hunt at end of course, one target microbe will be Pseudomonas, enrichment culture depends on its ability to grown anaerobically using nitrate reduction.
Sulfate (SO42-)
- Process called sulfate reduction.
- Sulfate (SO42-) Hydrogen Sulfide (H2S)
- Small group of bacteria carry out this reaction; all obligate anaerobes.
- Have unique cytochrome c3.
- Sulfate is common in sea water. Often, H2S combines with iron, forms insoluble FeS black sediments. Common in estuaries.
Carbon dioxide (CO2)
- One of most common inorganic ions.
- Methanogens: most important group of CO2 reducers. Obligate anaerobes, archaebacteria. Produce methane as waste product.
- Reaction: CO2 + H2 + H+ CH4 + H2O
- Note: reaction also requires Hydrogen gas. Methanogens typically live alongside bacteria that produce hydrogen by fermentation, remove hydrogen as it is made.
TCA cycle: further catabolism of pyruvate
Formation of acetyl-CoA
- Oxidation of pyruvate (3-C) + NAD+ Acetyl-CoA (2-C) + CO2 + NADH
- Carried out by pyruvate dehydrogenase (multi-enzyme system)
- Note: Acetyl-CoA can also be produced by breakdown of lipids or certain amino acids — important focal point of central metabolism
Net effects of TCA cycle:
- To start cycle:
- Acetyl-CoA (2-C) + oxaloacetate (4-C) citric acid (6-C)
- Subsequent steps:
- Convert citrate to isocitrate (still 6-C)
- Oxidize alpha-ketoglutarate (5-C) + CO2 + NADH
- Oxidize succinyl-CoA (4-C) + CO2 + NADH
- SLP reaction: succinyl-CoA (4-C) + GDP succinate (4-C) + GTP (Note: GTP can be interconverted with ADP to form ATP)
- Oxidize fumarate (4-C) + FADH2 — convert fumarate to malate
- (6)oxidize again oxaloacetate (4-C) + NADH
- Net yield: Acetyl-CoA (2-C) + 3 NAD+ + FAD 2 CO2 + 3 NADH + FADH2 + ATP
- TCA cycle completes the oxidation of carbons in pyruvate to most oxidized form (CO2); removes electrons originally in C-H bonds to electron carriers NADH and FADH for use in respiration machinery.
Catabolism of substances other than glucose: Many other possible C-sources for catabolism beside glucose. In general, must convert these into molecules that can enter into central metabolism, either in glycolysis or TCA cycle.
- carbohydrates
- Most abundant C-sources in most environments, most in various polysaccharides (cellulose, starch, lignin, etc.)
- To gain access to sugars, must first secrete hydrolytic enzymes that break down glycosidic bonds in polysaccharides, produce mono- and disaccharides that can be transported into cells.
- Starch, glycogen — easily hydrolyzed by amylases
- Cellulose — difficult to digest, very insoluble, tightly folded. Many fungi, some bacteria produce cellulases.
- Agar — some marine bacteria produce agars
- Once mono- or disaccharides are available, they are transported into cell, converted into some typical glycolytic intermediate such as glucose-6-phosphate, catabolized by glycolytic enzymes.
- lipids
- Biological lipids common as triglycerides, diglycerides.
- To catabolize, bacteria secrete lipases, hydrolyze glycerides to free fatty acids and glycerol.
- Fatty acids attacked by Beta-oxidation pathway.
- Using FAD and NAD+ to remove electrons, 2-C units are removed as Acetyl-CoA, feed directly into central metabolism at TCA cycle entry. Glycolysis pathway not involved (except for use in synthesizing sugars needed for cell wall, running sections of pathway in reverse).
- proteins
- Proteins must first be hydrolyzed by protease enzymes, to get individual amino acids which can be transported into cells.
- Amino acids all have common structure: NH2 – RCH – COOH.
- 1st step in catabolism is to remove amino group (deamination), often by swapping it with another substrate (transamination).
- Typical example: glutamic acid (an AA) + pyruvate alpha-ketoglutarate + alanine (= pyruvate + amino group). Now alpha-KG can be oxidized in TCA cycle, since it is a TCA cycle compound.
- As excess amino groups accumulate, must be secreted as waste products, possibly as ammonium ion (leads to alkaline pH).
- carbohydrates
|
| Download this lecture as PDF here |
Overview of Autotrophy
• Imagine being hungry, walking outside, taking off your shirt, lying in the sun for a few hours, becoming totally full (fat even!), and being done eating. No stores, no lines, no choices, just sunlight — and the machinery of an autotroph — and some CO2 and a couple of other requirements (water — and H2S or Hydrogen gas, if you happen to be an anerobe)
• Autotroph = gets all carbon from CO2, organic C not required (for C-source). Use special metabolic cycle: Calvin-Benson cycle
• Refers to C-source only; some organisms still require organic C as energy source
Calvin-Benson cycle
• Each CO2 is added to a 5-C acceptor molecule (ribulose 1,5 bis-phosphate)
• Immediately split into two 3-C molecules (3-phosphoglyceric acid)
• Must add phosphate group (from ATP) and hydrogen (from NADPH) to get reduced product, 3 – phospho-glyceraldehyde (PGA)
• Cannot take all (PGA) as product — must regenerate some more acceptor to keep cycle going. How?
• Take 5 PGA molecules (5 x 3C = 15 C atoms). Rearrange through series of reactions to make 3 5 – C molecules (still 15 C atoms). Add ATP to each, make 3 acceptor molecules (ribulose 1,5 bis-phosphate)
• Net result: To get 1 PGA (3-C) as reduced product, need 3 CO2 molecules, added to 3 acceptor molecules —-> six 3 – C molecules, use 6 ATP and 6 NADPH —-> 6 PGA molecules; five of these are used to regenerate acceptor molecules (+ 3ATP), one PGA can leave cycle and be used by cell.
Summary
• Actual cyce exports 3-C reduced molecules: look at balanced equation:
3 CO2 + 9 ATP + 6 NADPH —–> 3-phospho-glyceraldehyde (PGA) + 9 ADP + 9 NADP+
• Often want to look at balanced equation relative to 6C synthesis. Must multiply all terms in balanced equation above by two (since 2 PGA ~ 1 glucose)
6 CO2 + 18 ATP + 12 NADPH —–> glucose + 18 ADP + 12 NADP+
• Note for reaction: glucose + O2 —-> 6CO2 + 6 H2O; delta Go’= – 688kcal/mole
• If each ATP contains ~7.3 kcal/mole (from delta Go’ for hydrolysis) and each NADPH contains ~54 kcal/mole (from delta Go’ for oxidation), then to make glucose costs 780 kcal/mole, more than the energy available by oxidizing glucose.
• Conclusion: making sugar is expensive! Cell needs to supply large quantities of ATP and NADPH.
Chemolithotrophs
Hydrogen Bacteria
• Gain energy by oxidizing hydrogen gas:
• H2 + NAD+ ——-(hydrogenase enzyme)——> NADH + H+
• alternative: electrons can be donated directly to ETS chain, bypassing NAD
• Note: only need one special enzyme to carry this step out: hydrogenase.
• Many different genera of bacteria include members that can induce hydrogenase. When hydrogen disappears, back to heterotrophic life. Hydrogen bacteria are usually facultative chemolithotrophs.
Sulfur Bacteria
• Called “colorless” in contrast to chlorophyll-containing sulfur bacteria, usually green or purple
• Oxidize sulfur compounds: Example: Thiobacillus thiooxidans thiosulfate: S2O3= —–> SO4=free sulfur: 2 So + 2 H2O + 3 O2 —–> 2 H2SO4
• Note product: sulfuric acid!! Cells can grow even in pH 0 (1M sulfuric acid). But cell internal pH is ~7, so difference across membrane can be 6 or 7 pH units.
• Acid mine drainage: common in Western Penn., E. Ohio, W. Virginia. Rivers can run rust red. Mines have been major sources of pollution. Water seeps in, sulfur deposits exposed during coal mining allow microbial growth ——> megatons of H2SO4
• Sulfuric acid leaches out, dissolves iron, precipitates in river with bicarbonate to form rusty deposits.
• Quantities involved: Ohio River carries 100 million tons of 98% conc. H2SO4 per year.
• To cure problem, must seal up old mines, prevent oxygen access. Also strip mines must be promptly covered up once mining is done to block access of microbes and oxygen to sulfur.
• Value of this reaction:
(a) farmers or gardeners can dump free S on alkaline soil, bacteria will produce acid
(b) miners can use process to recover Cu from low grade ores, where smelting is not economical. Pile up mine “tailings” with copper ore; scrap shallow hole and fill with water. If tailings contain S, microbes will produce H2SO4. Now pump the acid over the tailings, Cu will be leached out and accumulate as soluble ions in acid pool. Eventually process the acid, recover Cu.
Nitrifying Bacteria
• Very important soil organisms — process all ammonia, nitrite in soils, break down amino acids, nitrogen bases —> ammonia (NH3)
• Two different groups: one oxidizes ammonia, one oxidizes nitrite
• Ex. 1: Nitrosomonas: 2 NH3 (ammonia) + 3 O2 —–> 2 HNO2 (nitrite) + 2 H2O
• Ex. 2: Nitrobacter: 2 HNO2 (nitrite) + 2 O2 —–> 2 HNO3 (nitrate)
• Note potential problem: redox potential for nitrite as electron donor is + 0.42 v., so can easily pass electrons down to oxygen at + 0.82 v., reaction will be spontaneous. Electrons can be passed through an electron transport system, make ATP by chemiosmotic phosphorylation.
• BUT — how to make NADPH? (Remember, this an autotroph, needs both ATP and NADPH to grow). How to get NADPH? The redox potential is much higher than nitrite.
• Solution: Reverse electron transport. Accumulate enough proton gradient by oxidation of nitrite to force electrons back to carriers with higher redox potentials, all the way back to NADH —> NADPH. This works as long as concentrations of reduced forms are kept very low, and NADPH is used up immediately to make glyceraldehyde-3-phosphate. See handout
• This is very inefficient process. Nitrobacter can have 18 hour generation time. But it has no competition, so what’s a little extra time?
Iron Bacteria
• Curious discovery: Ferrobacillus ferrooxidans. Carries out oxidation of iron: Fe++ (ferrous) —-> Fe+++ (ferric) + e-
• Originally thought bacteria get energy from oxidation, make ATP. But redox potential of Fe oxidation is + 0.78 v., and redox potential for oxygen is + 0.86 v., so delta Eo’ for aerobic respiration is only -0.08 v., calculated delta Go’ is much less than the 7.3 kcal/mole needed to make ATP. How does this organism grow?
• It only grows in very acidic habitats, pH less than 3. Found with Thiobacillus thiooxidans, bacterium that produces sulfuric acid. Ferrobacillus lives off the pH gradient created by acidic pH. This maintains very high proton gradient. As H+ flows in, ATP gets made. But need to get rid of H+ inside, keep internal pH at 7. Use Fe++ as electron donor to oxygen, combine with H+ to form water, get rid of outside cell. Iron functions as electron supplier to get rid of protons.
• Cells process an enormous amount of iron for very small yields of energy. Fe+++ reacts with OH- ions to form insoluble precipitate, Fe(OH)3, reddish yellow color.
Phototrophs
• Use energy from sunlight to get high energy electrons (attached to carriers high on redox tower). Use CO2 and Calvin-Benson cycle to make all organic molecules.
• Critical molecules: photon absorbers = bacteriochlorophylls. Several different varieties. Light is trapped by a patch of pigments = “antenna field”, gets passed around to a “reaction center” where an electron is released from Mg++ ion with high energy, passed to electron transport system — from this point, can use electron transport systems to generate proton gradients, make ATP.
• Problem: need to make not only ATP (available from proton gradient), but also NADPH. How to obtain?
• Two solutions:
use a reduced molecule with high redox potential like hydrogen gas (H2) or hydrogen sulfide (H2S) to pass electrons to NADP+. Light not needed for this.
use a reduced molecule with low redox potential like water to release electrons and H+ ions. Need lots of energy to drive this reaction, so need an extra step. Light is needed for this.
Anaerobic photosynthetic bacteria
• Three common groups:
Purple bacteria Exs: Chromatium vinosum, Thiospirillum jenense
Purple nonsulfur bacteria. Exs: Rhodospirillum rubrum, Rhodobacter sphaeroides vannielii
Green sulfur bacteria (many are actually brown) Exs: Chlorobium limicola, Prosthecochloris aestuarii,
• Notes: in both groups, electrons released by light travel through electron transport systems back to the original photosystem = cyclic electron flow. Proton gradient is produced, ATP is made as protons flow back through ATP synthase molecules. Specific carriers are different.
• To make NADPH, need reduced electron donor. (1) in purple bacteria, can use organic molecules (e.g. fumarate), or H2 for non-sulfur bacteria; or can use H2S or H2 for purple sulfur bacteria. Sulfur accumulates inside cells when H2S is used, hence the name. (2) in green sulfur bacteria, can use H2S, or H2. Sulfur accumulates outside cells.
Aerobic photosynthetic bacteria = cyanobacteria
• includes both prokaryotes (cyanobacteria, formerly called blue-green bacteria) and eukaryotes (algae, green plants)
• Two photosystems are needed, not one as in anoxygenic photosynthesis. Why?
• Source of NADPH = electrons removed from photosystem I —> excited by light to high redox potential, passed to ferredoxin, then directly to NADP+ —–> NADPH
• Now photosystem I has + charge, can’t supply any more electrons. Can’t have this, so replace electrons from another photosystem II (see handout diagram), also energized by light. During this process, electrons flow through ETS system and make a proton gradient ( ——> ATP by chemiosmotic phosphorylation). But electrons aren’t flowing back to same place they started from — this is non-cyclic electron flow. Path resembles a letter “Z”, so often called “Z-scheme” photosynthesis.
FERMENTATION
• Fermentation — oxidation of an organic compound in the absence of external electron acceptor (no oxygen required). Uses SLP (substrate-level phosphorylation).
• Respiration — oxidation of an organic compound where oxygen is the final electron acceptor. Uses ETS (electron transport system) as well as SLP.
• Anaerobic respiration (unique to bacteria) — oxidation of organic compounds where an external substrate other than oxygen serves as final electron acceptor. Exs: nitrate, sulfate, carbon dioxide.
Lactic acid fermentation
• pyruvate + NADH lactic acid + NAD+
• found in many bacteria: lactic acid bacteria, Bacillus, also in some protozoa, water molds, even human skeletal muscle
• Responsible for souring of milk products yogurt, cheese, buttermilk, sour cream, etc. Excellent keeping properties.
• Some bacteria produce only lactic acid = Homolactic fermenters
• Other bacteria produce other products as well; ethanol, CO2, lactate, etc. = Heterolactic fermenters
Alcoholic fermentation
• pyruvate acetaldehyde + CO2
• acetaldehyde + NADH ethanol + NAD+
• Found in many fungi, yeasts, some bacteria.
• Very important in human applications. Bread, alcoholic spirits
Formic acid and mixed acid fermentations
• pyruvate (3-C) + CoA Acetyl-CoA (2-C) + formic acid (1-C)
• HCOOH CO2 + H2
• found in many bacteria, very common in enterics (Gram-negative facultative anaerobic rods, include E. coli and other common intestinal tract denizens)
Useful in identification: 2 common variants
Mixed acid fermentation: Some bacteria use several pathways, produce ethanol, formic acid, acetic acid, lactic acid, succinic acid, CO2, and H2. Note lots of acid, lower pH than many other fermentations. Note: ATP yield via mixed acid is ~2.5 ATP/glucose, a bit higher than straight lactic acid fermentation
Butanediol fermentation: Butanediol produced, also much more CO2, and H2
Roles of fermentation in nature
• Fermentations play major role.
• large part of cellulose ingested by herbivores is excreted in undigested form.
• Wherever organic matter accumulates, bacteria can grow and remove oxygen (by respiration), leading to anaerobic conditions that favor fermentation.
• Even in lab cultures (test tubes of media), bacteria eat up all available oxygen, rely largely on fermentation unless vigorous aeration is maintained! Bacteria are pigs, gorge themselves at every opportunity!
• Beside bacteria, fermentations also carried out be protozoa, fungi, even animal muscle tissues (only works as temporary energy supplement).
What substances can be fermented?
• must have intermediate oxidation state (o.s.)
• if totally oxidized (-CO)n cannot be fermented
• if totally reduced (-CH2)n, cannot be fermented
• must be convertible to a substrate for substrate level phosphorylation (usually into some glycolytic step)
• Many sugars can be fermented. Also amino acids (e.g. by Clostridia, oxidizing one amino acid and using a different amino acid as electron acceptor.)
Respiration
• Use an external electron acceptor. Oxygen as prototype.
• The “problem” with fermentation is that, by using an organic molecule as a terminal electron acceptor to be discarded as waste, cell is losing out on potential to further oxidize organic molecule, get more energy.
• Alternative solution is to use some non-organic molecule that has a low redox potential, can accept electrons and become some reduced molecule. Oxygen is perfect for this, has extremely low redox potential, and becomes reduced to water, the “perfect” waste product for an aqueous environment.
• To transfer electrons (and protons, H+) to oxygen, need special oxidase enzyme. In mitochondria, this is a cytochrome, cyt a. In bacteria, different cytochromes; in E. coli, cyt o or d.
| Download this lecture as PDF here |
Bacteriophage (phage) are obligate intracellular parasites that multiply inside bacteria by making use of some or all of the host biosynthetic machinery (pathmicro.med.sc.edu/ppt). The term is commonly used in its shortened form, phage. Interestingly, Bacteriophages are much smaller than the bacteria they destroy.Phages are estimated to be the most widely distributed and diverse entities in the biosphere. Phages are ubiquitous and can be found in all reservoirs populated by bacterial hosts, such as soil or the intestines of animals. One of the densest natural sources for phages and other viruses is sea water. They have been used for over 60 years as an alternative to antibiotics, however, this much controversial area of research (http://www.phages.org).
Typical phages have hollow heads (where the phage DNA or RNA is stored) and tunnel tails, the tips of which have the ability to bind to specific molecules on the surface of their target bacteria. The viral DNA is then injected through the tail into the host cell, where it directs the production of progeny phages often over a hundred in half an hour. These “young” phages burst from the host cell (killing it) and infect more bacteria.

Composition of bacteriophages
Although different bacteriophages may contain different materials they all contain nucleic acid and protein. Depending upon the phage, the nucleic acid can be either DNA or RNA but not both and it can exist in various forms. Bacteriophages have been classified as:
The nucleic acids of phages often contain unusual or modified bases. These modified bases protect phage nucleic acid from nucleases that break down host nucleic acids during phage infection. The size of the nucleic acid varies depending upon the phage. The simplest phages only have enough nucleic acid to code for 3-5 average size gene products while the more complex phages may code for over 100 gene products.
The number of different kinds of protein and the amount of each kind of protein in the phage particle will vary depending upon the phage. The simplest phages have many copies of only one or two different proteins while more complex phages may have many different kinds. The proteins function in infection and to protect the nucleic acid from nucleases in the environment. Phages are also commonly employed in gene cloning, especially those exhibiting lytic and lysogenic cycles (http://www.web-books.com)
Structure of bacteriophages
Bacteriophage comes in many different sizes and shapes. The basic structural features of bacteriophages are (which depicts the phage called T4)
1. Size – T4 is among the largest phages; it is approximately 200 nm long and 80-100 nm wide. Other phages are smaller. Most phages range in size from 24-200 nm in length.
2. Head or Capsid – All phages contain a head structure which can vary in size and shape. Some are icosahedral (20 sides) others are filamentous. The head or capsid is composed of many copies of one or more different proteins. Inside the head is found the nucleic acid. The head acts as the protective covering for the nucleic acid.
3. Tail – Many but not all phages have tails attached to the phage head. The tail is a hollow tube through which the nucleic acid passes during infection. The size of the tail can vary and some phages do not even have a tail structure. In the more complex phages like T4 the tail is surrounded by a contractile sheath which contracts during infection of the bacterium. At the end of the tail the more complex phages like T4 have a base plate and one or more tail fibers attached to it. The base plate and tail fibers are involved in the binding of the phage to the bacterial cell. Not all phages have base plates and tail fibers. In these instances other structures are involved in binding of the phage particle to the bacterium.

Infection of Host Cells (http:// biology.about.com/od/virology)

A. Adsorption
The first step in the infection process is the adsorption of the phage to the bacterial cell. This step is mediated by the tail fibers or by some analogous structure on those phages that lack tail fibers and it is reversible. The tail fibers attach to specific receptors on the bacterial cell and the host specificity of the phage (i.e. the bacteria that it is able to infect) is usually determined by the type of tail fibers that a phage has. The nature of the bacterial receptor varies for different bacteria. Examples include proteins on the outer surface of the bacterium, LPS, pili, and lipoprotein. These receptors are on the bacteria for other purposes and phages have evolved to use these receptors for infection.
B. Irreversible attachment
The attachment of the phage to the bacterium via the tail fibers is a weak one and is reversible. Irreversible binding of phage to a bacterium is mediated by one or more of the components of the base plate. Phages lacking base plates have other ways of becoming tightly bound to the bacterial cell.VIE
C. Sheath Contraction
The irreversible binding of the phage to the bacterium results in the contraction of the sheath (for those phages which have a sheath) and the hollow tail fiber is pushed through the bacterial envelope. Phages that don’t have contractile sheaths use other mechanisms to get the phage particle through the bacterial envelope. Some phages have enzymes that digest various components of the bacterial envelope.
D. Nucleic Acid Injection
When the phage has gotten through the bacterial envelope the nucleic acid from the head passes through the hollow tail and enters the bacterial cell. Usually, the only phage component that actually enters the cell is the nucleic acid. The remainder of the phage remains on the outside of the bacterium. There are some exceptions to this rule. This is different from animal cell viruses in which most of the virus particle usually gets into the cell. This difference is probably due to the inability of bacteria to engulf materials.

LYTIC AND LYSOGENIC CYCLES – PHAGE MULTIPLICATION CYCLE
A. Definition – Lytic or virulent phages are phages which can only multiply on bacteria and kill the cell by lysis at the end of the life cycle.


Lytic or Virulent Phages
a. Eclipse period – During the eclipse phase, no infectious phage particles can be found either inside or outside the bacterial cell. The phage nucleic acid takes over the host biosynthetic machinery and phage specified m-RNA’s and proteins are made. There is an orderly expression of phage directed macromolecular synthesis, just as one sees in animal virus infections. Early m-RNA’s code for early proteins which are needed for phage DNA synthesis and for shutting off host DNA, RNA and protein biosynthesis. In some cases the early proteins actually degrade the host chromosome. After phage DNA is made late m-RNA’s and late proteins are made. The late proteins are the structural proteins that comprise the phage as well as the proteins needed for lysis of the bacterial cell.
b. Intracellular Accumulation Phase – In this phase the nucleic acid and structural proteins that have been made are assembled and infectious phage particles accumulate within the cell.
c. Lysis and Release Phase – After a while the bacteria begin to lyse due to the accumulation of the phage lysis protein and intracellular phage are released into the medium. The number of particles released per infected bacteria may be as high as 1000.
Assay for Lytic Phage
a. Plaque assay – Lytic phage are enumerated by a plaque assay. A plaque is a clear area which results from the lysis of bacteria. Each plaque arises from a single infectious phage. The infectious particle that gives rise to a plaque is called a pfu (plaque forming unit).
B. Lysogenic or Temperate Phage
1. Definition – Lysogenic or temperate phages are those that can either multiply via the lytic cycle or enter a quiescent state in the cell. In this quiescent state most of the phage genes are not transcribed; the phage genome exists in a repressed state. The phage DNA in this repressed state is called a prophage because it is not a phage but it has the potential to produce phage. In most cases the phage DNA actually integrates into the host chromosome and is replicated along with the host chromosome and passed on to the daughter cells. The cell harboring a prophage is not adversely affected by the presence of the prophage and the lysogenic state may persist indefinitely. The cell harboring a prophage is termed a lysogen.
2. Events Leading to Lysogeny – The Prototype Phage: Lambda
a. Circularization of the phage chromosome – Lambda DNA is a double stranded linear molecule with small single stranded regions at the 5′ ends. These single stranded ends are complementary (cohesive ends) so that they can base pair and produce a circular molecule. In the cell the free ends of the circle can be ligated to form a covalently closed circle as illustrated in Figure 5.
b. Site-specific recombination – A recombination event, catalyzed by a phage coded enzyme, occurs between a particular site on the circularized phage DNA and a particular site on the host chromosome. The result is the integration of the phage DNA into the host chromosome as illustrated in Figure 6.
c. Repression of the phage genome – A phage coded protein, called a repressor, is made which binds to a particular site on the phage DNA, called the operator, and shuts off transcription of most phage genes EXCEPT the repressor gene. The result is a stable repressed phage genome which is integrated into the host chromosome. Each temperate phage will only repress its own DNA and not that from other phage, so that repression is very specific (immunity to superinfection with the same phage).
3. Events Leading to Termination of Lysogeny
Anytime a lysogenic bacterium is exposed to adverse conditions, the lysogenic state can be terminated. This process is called induction. Conditions which favor the termination of the lysogenic state include: desiccation, exposure to UV or ionizing radiation, exposure to mutagenic chemicals, etc. Adverse conditions lead to the production of proteases (rec A protein) which destroy the repressor protein. This in turn leads to the expression of the phage genes, reversal of the integration process and lytic multiplication.

4. Lytic vs Lysogenic Cycle
The decision for lambda to enter the lytic or lysogenic cycle when it first enters a cell is determined by the concentration of the repressor and another phage protein called cro in the cell. The cro protein turns off the synthesis of the repressor and thus prevents the establishment of lysogeny. Environmental conditions that favor the production of cro will lead to the lytic cycle while those that favor the production of the repressor will favor lysogeny.
5. Significance of Lysogeny
a. Model for animal virus transformation – Lysogeny is a model system for virus transformation of animal cells
b. Lysogenic conversion – When a cell becomes lysogenized, occasionally extra genes carried by the phage get expressed in the cell. These genes can change the properties of the bacterial cell. This process is called lysogenic or phage conversion. This can be of significance clinically. e.g. Lysogenic phages have been shown to carry genes that can modify the Salmonella O antigen, which is one of the major antigens to which the immune response is directed. Toxin production by Corynebacterium diphtheriae is mediated by a gene carried by a phage. Only those strain that have been converted by lysogeny are pathogenic.
| Download this lecture as PDF here |
A. Definition – Lytic or virulent phages are phages which can only multiply on bacteria and kill the cell by lysis at the end of the life cycle.
Lytic or Virulent Phages
a. Eclipse period – During the eclipse phase, no infectious phage particles can be found either inside or outside the bacterial cell. The phage nucleic acid takes over the host biosynthetic machinery and phage specified m-RNA’s and proteins are made. There is an orderly expression of phage directed macromolecular synthesis, just as one sees in animal virus infections. Early m-RNA’s code for early proteins which are needed for phage DNA synthesis and for shutting off host DNA, RNA and protein biosynthesis. In some cases the early proteins actually degrade the host chromosome. After phage DNA is made late m-RNA’s and late proteins are made. The late proteins are the structural proteins that comprise the phage as well as the proteins needed for lysis of the bacterial cell.
b. Intracellular Accumulation Phase – In this phase the nucleic acid and structural proteins that have been made are assembled and infectious phage particles accumulate within the cell.
c. Lysis and Release Phase – After a while the bacteria begin to lyse due to the accumulation of the phage lysis protein and intracellular phage are released into the medium. The number of particles released per infected bacteria may be as high as 1000.
Assay for Lytic Phage
a. Plaque assay – Lytic phage are enumerated by a plaque assay. A plaque is a clear area which results from the lysis of bacteria. Each plaque arises from a single infectious phage. The infectious particle that gives rise to a plaque is called a pfu (plaque forming unit).
B. Lysogenic or Temperate Phage
1. Definition – Lysogenic or temperate phages are those that can either multiply via the lytic cycle or enter a quiescent state in the cell. In this quiescent state most of the phage genes are not transcribed; the phage genome exists in a repressed state. The phage DNA in this repressed state is called a prophage because it is not a phage but it has the potential to produce phage. In most cases the phage DNA actually integrates into the host chromosome and is replicated along with the host chromosome and passed on to the daughter cells. The cell harboring a prophage is not adversely affected by the presence of the prophage and the lysogenic state may persist indefinitely. The cell harboring a prophage is termed a lysogen.
2. Events Leading to Lysogeny – The Prototype Phage: Lambda
a. Circularization of the phage chromosome – Lambda DNA is a double stranded linear molecule with small single stranded regions at the 5′ ends. These single stranded ends are complementary (cohesive ends) so that they can base pair and produce a circular molecule. In the cell the free ends of the circle can be ligated to form a covalently closed circle as illustrated in Figure 5.
b. Site-specific recombination – A recombination event, catalyzed by a phage coded enzyme, occurs between a particular site on the circularized phage DNA and a particular site on the host chromosome. The result is the integration of the phage DNA into the host chromosome as illustrated in Figure 6.
c. Repression of the phage genome – A phage coded protein, called a repressor, is made which binds to a particular site on the phage DNA, called the operator, and shuts off transcription of most phage genes EXCEPT the repressor gene. The result is a stable repressed phage genome which is integrated into the host chromosome. Each temperate phage will only repress its own DNA and not that from other phage, so that repression is very specific (immunity to superinfection with the same phage).
3. Events Leading to Termination of Lysogeny
Anytime a lysogenic bacterium is exposed to adverse conditions, the lysogenic state can be terminated. This process is called induction. Conditions which favor the termination of the lysogenic state include: desiccation, exposure to UV or ionizing radiation, exposure to mutagenic chemicals, etc. Adverse conditions lead to the production of proteases (rec A protein) which destroy the repressor protein. This in turn leads to the expression of the phage genes, reversal of the integration process and lytic multiplication.
4. Lytic vs Lysogenic Cycle
The decision for lambda to enter the lytic or lysogenic cycle when it first enters a cell is determined by the concentration of the repressor and another phage protein called cro in the cell. The cro protein turns off the synthesis of the repressor and thus prevents the establishment of lysogeny. Environmental conditions that favor the production of cro will lead to the lytic cycle while those that favor the production of the repressor will favor lysogeny.
5. Significance of Lysogeny
a. Model for animal virus transformation – Lysogeny is a model system for virus transformation of animal cells
b. Lysogenic conversion – When a cell becomes lysogenized, occasionally extra genes carried by the phage get expressed in the cell. These genes can change the properties of the bacterial cell. This process is called lysogenic or phage conversion. This can be of significance clinically. e.g. Lysogenic phages have been shown to carry genes that can modify the Salmonella O antigen, which is one of the major antigens to which the immune response is directed. Toxin production by Corynebacterium diphtheriae is mediated by a gene carried by a phage. Only those strain that have been converted by lysogeny are pathogenic.
| Download this lecture as PDF here |
A virus is a small infectious agent that can replicate only inside the living cells of organisms. Most viruses are too small to be seen directly with a light microscope. Viruses infect all types of organisms, from animals and plants to bacteria and archaea. Since the initial discovery of tobacco mosaic virus by Martinus Beijerinck in 1898, about 5,000 viruses have been described in detail though there are millions of different types. Viruses are found in almost every ecosystem on Earth and are the most abundant type of biological entity. The study of viruses is known as virology, a sub-speciality of microbiology.
Virus particles (known as virions) consist of two or three parts: the genetic material made from either DNA or RNA, long molecules that carry genetic information; a protein coat that protects these genes; and in some cases an envelope of lipids that surrounds the protein coat when they are outside a cell. The shapes of viruses range from simple helical and icosahedral forms to more complex structures. The average virus is about one one-hundredth the size of the average bacterium.
- Every virus has 2 stages
- dormant, particulate, transmissible stage called the virion stage
- an active, intracellular stage called the infectious stage
Virion Stage
- Virions are the transmissible state of a virus. Metabolically inert.
- Virion = “a piece of nucleic acid wrapped up in a protein coat” (and/or a membrane)
- The nucleic acid can be either DNA (double-stranded (ds) or single-stranded (ss)) or RNA (ds or ss); never both.
- The coat (also called viral shell or capsid) can be icosahedron (20-sided regular geometric shape common in many bacterial, animal, and plant viruses), sphere, cylinder, bullet-shaped, or amorphous shaped particle.
- Virions must be able to adhere and allow entry into some host cell(s). Also to survive outside of host cell environment.
- Some virions more hardy than others (e.g., hepatitis virus can withstand short periods of boiling; most virions are destroyed by this).
Infectious Stage
- When virus infects a cell, nucleic acid must be uncoated and gain access to metabolic machinery of cell.
- Virus life cycle is characterized by:
- attachment
- penetration, with entry of nucleic acid into cell
- early expression of virus genes (either directly by translation, if virus contains “+” RNA, or indirectly after transription and then translation)
- replication of virus nucleic acid
- synthesis of new virion components
- packaging and assembly of new virions
- exit from cell
Measurement of viral growth
- Must grow virus on host cells to see anything. Can’t grow virus without cells.
- To quantify viruses, need some way to get flat surface of growing cells, allow virus-infected cells to spread radially where present = plaque.
- In bacterial cells this is easy. Spread “lawn” of bacteria on plate, add diluted phage suspension or culture infected with phages. After 6-8 hours can see plaques in E. coli.
- In plant cells, can be easy. Example: Tobacco Mosaic Virus (TMV), make virus dilution, rub over surface of tobacco leaf. After leaf growth, can observe plaque areas.
- In animal cells, not so easy. In 1960’s, standard assay was to inoculate chicken egg membranes of developing chick embryos, incubate for a week, cut open shell and count plaques on membrane in the air sac. Lots of work to get statistically reliable data!
- In 1970’s tissue culture became a viable alternative. Animal cells are cultured as microbes in glass or plastic, use special medium that contains most of nutrients present in blood. Cells will spread as monolayer on surface, can count plaques after staining.
Taxonomy of viruses
- Based mainly on Virion and Kingdom of host
- Use Host cell type (Animal viruses, plant viruses, etc.)
- Use Nucleic Acid type (ds DNA, ss DNA, ds RNA, ss RNA)
- Use + or – polarity of RNA. “+” is able to serve as mRNA. “-” is the complement of +, must function as template to make a complementary strand of + RNA before any translation can occur.
- Use virus coat morphology. Enveloped vs. non-enveloped viruses.
Virion Structure
“Naked” viruses
- Helical viruses
- Tobacco mosaic virus (TMV) is an example of a virus with helical symmetry.
- A helical array of identical protein subunits surrounds an RNA molecule.
- Helical viruses

- Icosahedral viruses
- built from icosahedral (20-sided) assemblies of protein subunits.
- Icosahedral shape is the minimum free energy structure for producing a shell of equivalently bonded identical structures.
- The simplest icosahedral capsids are built up by using 3 identical subunits to form each triangular face, thereby requiring 60 identical subunits to form a complete capsid. A few simple virus particles are constructed in this way, e.g. bacteriophage ØX174.
- Most icosahedral viruses have more than 60 subunits, usually some multiple N times 60. N (called the triangulation number) can have values of 1, 3, 4, 7, 9, 12, and more.

- Icosahedral viruses
“Enveloped” viruses
- “Naked” viruses require host death so viruses can be released. This may be wasteful, and may cause premature death of host cell.
- Alternative strategy: shed virus particles by budding out, continued release from cell membrane. Cell does not die (immediately), continues to serve as factory for virus assembly and release. Virus typically acquires a coating of host cell membrane, modified to include virus-specific proteins. This is the “envelope”. Virus may have additional protein coats (often icosahedral) inside the envelope.
- Eventually host cell is too depleted to survive. Can see evidence of this as “cytopathic effect” (CPE). Cell then dies.
- Examples of enveloped viruses include:
- Retrovirus, including HIV
- Paramyxovirus, including influenza
- Rhabdovirus, including rabies
- Filovirus. Although very “hot” in the news, these viruses are very poorly characterized because of their extreme pathogenicity. They are class IV pathogens, meaning they can only be cultured in total containment facilities, of which there are only two in the U. S. They are thought to be enveloped viruses with – RNA genomes.
Virus Genomes
- Rule of Thumb: to estimate # of virus proteins, look at size of viral DNA or RNA. For each 1000 base pairs, can guess the existence of 1 protein
- “typical” gene has 300-400 amino acids = ~ 1000 base pairs = 1 kbp (= 1 protein)
- small virus: SV40 => 5000 base pairs = 5 kbp ~ 5 proteins
- large virus: T4 => 200 kbp ~ 100-200 proteins
- by comparison, E. coli: 4000 kbp
Bacterial Viruses = Phages
Bacterial defenses against infection
Cell surfaces: possibilities of mutation
- Virus must attach to some specific cell surface protein or polysaccharide. But these are specified by genes, and genes can mutate. In population, will always find some variant strains with slightly different cell surfaces, may not bind virus well.
- When phage first discovered, thought this could be effective weapon against bacterial disease. But frequency of resistant bacterial strains was too high, any given strain of virus quickly became useless as resistant survivors propagated.
Nucleases: endo- and exo-DNases and RNases
- All bacteria seem to have nucleases that can attack DNA (called DNases) and RNA (called RNases).
- Exoenzymes attack free 5′ or 3′ ends of DNA, RNA molecules. Bacteria are protected since DNA (and plasmids) are always circular. RNases are present, and in fact destroy mRNA eventually (bacteria are always making new RNAs, very responsive to enviroment changes).
- Endonucleases are potentially lethal weapons. Called restriction enzymes. Attack at specific sequence: e.g., in E. coli, enzyme called EcoRI will attack any sequence with 5′ G-A-A-T-T-C 3′ (cuts DNA between G and A).
- Why doesn’t this kill cell? Because cell also has a second enzyme, called modification enzyme, that protects all host DNA sequences of this type. Typically adds a methyl (-CH3) group to one base at the cutting site. The methylated base is modified, and protected from the restriction enzyme. When foreign DNA comes into cell (e.g. virus DNA), if restriction site if present it will be cut and —– requiem for the virus.
The importance of Restriction Enzymes
Restriction enzymes are responsible for the genetic revolution. They make reproducible, specific cuts with surgical precision. Major industry has emerged in biochemical supply companies to harvest bacteria, purify restriction enzymes, and sell these to research and applied industries.
Animal Viruses
- Animal viruses are different in many respects from bacterial viruses. The host cells are more complex, with multiple compartments and more complex regulation of replication, transcription, and translation. Animal cells are not bounded by cell walls.
- Not surprisingly, animal viruses have evolved to overcome these problems. They attach and enter by different mechanisms than phages, and their intracellular activities include the ability to move between different compartments as needed.
- Viral entry and exit from cells is very different from bacteriophages. Animal viruses must enter through cell membrane, either by triggering endocytosis pathway or by fusing viral envelope with the cell envelope.
- Modifications are needed in both cellular and viral mRNA to allow recognition and movement from nucleus to cytoplasm. For example:
- 3′ tail of poly A
- 5′ cap of methyl Guanosine triphosphate

Types of infection
Viruses in animal cells show a variety of infection patterns:
- lytic infection: destroys host cells.
- persistent infection: host cell continues to shed virus over long time. Cell gradually becomes recognizably poorer (recognized as cytopathic effect, or CPE), eventually “crumps out”.
- transformation: infection by certain viruses causes cells to change, become cancerous. Responsible genes are called oncogenes (tumor-producing genes). Viral oncogenes have also been found in uninfected cells. These are genes involved in regulation of cell cycle; when defective, normal regulatory control is lost and cell can become cancerous.
- latent infection: virus genes may not be expressed for long time (ex. many Herpes infections). Not the same as lysogeny — genes are not integrated into host chromosome.
Human Immunodefiency Virus (HIV) and AIDS
- AIDS first recognized in 1981. Over 300,000 cases reported in U.S., over 8 million in Africa, over 12 million infected world-wide.
- View AIDS in perspective: a PBS website with current data on the AIDS epidemic.
- Transmission: sexual activity, especially with multiple sex partners. Also contaminated blood, needles, hospitals. Not just a disease of homosexuals! In Africa (most # cases) about equal # of male and female victims.
- AIDS lowers immune system’s ability to respond to other infections, allows opportunistic pathogens to invade body. Most common infection is pneumonia (lung infection) caused by Pneumocystis (2/3 of all AIDS patients get this at some point).
- Host cell for the virus is CD4 (T-helper) cell, needed to activate antibody production. In normal human, CD4 cells account for 70% of total T cells — in AIDS, number decreases, may reach 0% of T cell pool.
- Progress of HIV infection only recently understood. Formerly thought that virus became latent. Now discover that virus is anythig but latent: during infected period (which can last 10 years), body is destroying ~ billion virions/day, and virus is killing about 100 million CD4 T cells a day. HIV virus continues replicating, and body rapidly replenishes lost T cells. Only when lymph nodes wear out does virus gain the upper hand. See handout in class titled “Huge HIV turnover helps explain drug resistance, pathogenicity”.
- Prognosis: with carefully selected treatments, better than before. Virtually every infected person dies sooner of later, usually within 10 years of infection. No cure known, no vaccine yet available. Virus mutates rapidly, many strain variations. Vaccines being tried, results mixed but preliminary.
- Drugs: some types of drugs offer limited success.
- AZT (azidothymidine) is analog of thymidine, but is blocked at 3′-position, so no further chain growth possible. These target viral reverse transcriptase enzyme. Should reduce DNA synthesis in treated cells. But eventually, viral mutants resistant to drug arise. Also, long term use of drug can cause toxic side effects.
- protease inhibitors. Like many viruses, HIV needs to cleave large protein product into smaller products, using viral protease protein. By inhibiting this enzyme, should block necessary stage in viral replication cycle. Still under development, but resistant viral mutants to these type of drugs have already been found. Still, drug offers promise. See handout article for more details. Safe sex! Caution with sharps. Extra caution in clinical settings.
HIV Life Cycle
(budding through plasma membrane)
Viroids and Prions
- Viroids very small ss RNA genomes (~300 nucleotides). No coat, and RNA does not encode protein. Known viroids cause diseases in plants because host cells replicate the RNA.
- Prions (protein infectious agent) do not have a nucleic acid genome. Prion diseases are often called spongiform encephalopathies because of the post mortem appearance of the brain with large vacuoles in the cortex and cerebellum.
- Pathology of brains infected with prion diseases:
- Scrapie (sheep)
- bovine spongiform encephalopathy (cows) = “mad cow disease”
- Creutzfeldt-Jakob Disease (humans)
- Prion diseases in humans are probably primarily a genetic neurotoxic disorder. Transmission of the disease to humans via infectious prions is likely to be rare.
- The prion is a modified form of a normal cellular protein known as PrPc (for cellular), found predominantly on the surface of neurons and thought to be involved in synaptic function.
The modified form of PrPc (= prion) is known as PrPsc (for scrapie) which is relatively resistant to proteases and accumulates in cytoplasmic vesicles of diseased individuals. Prion protein may cause normal protein to fold abnormally.
| Download this lecture as PDF here |
Bacterial genetics is the study of gene structure and function in bacteria. Genetics itself is concerned with determining the number, location, and character of the genes of an organism. The process of replication is essential to understand in or manipulate the genome and to understand the functioning of these organisms.
DNA REPLICATION
In general, DNA is replicated by uncoiling of the helix, strand separation by breaking of the hydrogen bonds between the complementary strands, and synthesis of two new strands by complementary base pairing. Replication begins at a specific site in the DNA called the origin of replication.
How does the DNA in the bacterial cell replicate ……
Transcription in bacteria… how does it happen……
Transcription is the process by which genetic information from DNA is transferred into RNA. DNA sequence is enzymatically copied by RNA polymerase to produce a complementary nucleotide RNA strand. One significant difference between RNA and DNA sequence is the presence of U, or uracil in RNA instead of the T, or thymine of DNA. In the case of protein-encoding DNA, transcription is the first step that ultimately leads to the translation of the genetic code, via the mRNA intermediate, into a functional peptide or protein. The stretch of DNA that is transcribed into an RNA molecule is called a transcription unit. A transcription unit that is translated into protein contains sequence that directs and regulates protein synthesis in addition to coding sequence that is translated into protein. Regulatory sequence that is before, or 5′, of the coding sequence is called 5′ untranslated (5’UTR) sequence, and sequence found following, or 3′, of the coding sequence is called 3′ untranslated (3’UTR) sequence. As in DNA replication, transcription proceeds in the 5′ → 3′ direction. The DNA template strand is read 3′ → 5′ by RNA polymerase and the new RNA strand is synthesized in the 5’→ 3′ direction. RNA polymerase binds to the 3′ end of a gene (promoter) on the DNA template strand and travels toward the 5′ end. Except for the fact that thymines in DNA are converted to uracils in RNA, the newly synthesized RNA strand will have the same sequence as the coding (non-template) strand of the DNA.
Prokaryote Eukaryote

How to undertake experiments for understanding genetics of bacteria ?
The classical way to investigate genes is to mate two organisms with different genotypes and compare the observable properties (phenotypes) of the parents with those of the progeny. Bacteria do not mate (in the usual way), so there is no way of getting all the chromosomes of two different bacteria into the same cell. However, there are a number of ways in which a part of the chromosome or genome from one bacterium can be inserted into another bacterium so that the outcome can be studied.
The first step in performing genetic research on bacteria is to select mutants that differ from wild-type cells in one or more genes. Then crosses are made between mutants and wild types, or between two different mutants, to determine dominance-recessive relationships, chromosomal location, and other properties. Various genetic methods are used to select bacterial mutants, antibiotic-resistant cells, cells with specific growth requirements, and so on.
- Mutants in bacteria are mostly biochemical in nature, because we can’t generally see the cells.
- The most important mutants are auxotrophs. An auxotroph needs some nutrient that the wild type strain (prototroph) can make for itself. For example, a trp- auxotroph can’t make its own tryptophan (an amino acid). To grow trp- bacteria, you need to add tryptophan to the growth medium. Prototrophs are trp+; they don’t need any tryptophan supplied since they make their own.
- Chemoauxotrophs are mutants that can’t use some nutrient (usually a sugar) that prototrophs can use as food. For example, lac- mutants can’t grow on lactose (milk sugar), but lac+ prototrophs can grow on lactose.
- Resistance mutants confer resistance to some environmental toxin: drugs, heavy metals, bacteriophages, etc. For instance, AmpR causes bacteria to be resistant to ampicillin, a common antibiotic related to penicillin.
- Auxotrophs and chemoauxotrophs are usually recessive; drug resistance mutants are usually dominant. A common way to find bacterial mutants is replica plating, which means making two identical copies of the colonies on a petri plate under different conditions.
- For instance, if you were looking for trp- auxotrophs, one plate would contain added tryptophan and the other plate would not have any tryptophan in it.
- Bacteria are first spread on the permissive plate, the plate that allows both mutants and wild type to grow, the plate containing tryptophan in this case. They are allowed to grow fro a while, then a copy of the plate is made by pressing a piece of velvet onto the surface of the plate, then moving it to a fresh plate with the restrictive condition (no tryptophan). The velvet transfers some cells from each colony to an identical position on the restrictive plate.
- Colonies that grow on the permissive plate but not the restrictive plate are (probably) trp- auxotrophs, because they can only grow if tryptophan is supplied.
Replica plating 
The Ames test uses bacteria to test chemicalsfor capacity to cause mutations, as well as carcinogens (cancer-causing chemicals). The procedure is as fiollows:


| Download this lecture as PDF here |
Is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as rRNA genes or tRNA genes, the product is a functional RNA. The process of gene expression is used by all known life – eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea) and viruses – to generate the macromolecular machinery for life. Several steps in the gene expression process may be modulated, including the transcription, RNA splicing, translation, and post-translational modification of a protein. Gene regulation gives the cell control over structure and function, and is the basis for cellular differentiation, morphogenesis and the versatility and adaptability of any organism. Gene regulation may also serve as a substrate for evolutionary change, since control of the timing, location, and amount of gene expression can have a profound effect on the functions (actions) of the gene in a cell or in a multicellular organism. In genetics, gene expression is the most fundamental level at which genotype gives rise to the phenotype. The genetic code is “interpreted” by gene expression, and the properties of the expression products give rise to the organism’s phenotype.
In genetics, gene expression is the most fundamental level at which genotype gives rise to the phenotype. The genetic code is “interpreted” by gene expression, and the properties of the expression products give rise to the organism’s phenotype.
Transcription (Video)
The gene itself is typically a long stretch of DNA. It is a blueprint for the production of RNA. The production of RNA copies of the DNA is called transcription, and is performed by RNA polymerase, which adds one RNA nucleotide at a time to a growing RNA strand. This RNA is complementary to the template 3′ → 5′ DNA strand,[1] which is itself complementary to the coding 5′ → 3′ DNA strand. Therefore, the resulting 5′ → 3′ RNA strand is identical to the coding DNA strand with the exception that thymines are replaced with uracils in the RNA. A coding DNA strand reading “ATG” is transcribed as “UAC” in RNA.
- Initiation
- 30S initiates binding to mRNA.
- locates Shine-Dalgarno sequence (3-9 bases near 5′ end of mRNA).
- ribosome finds first AUG codon.
- 50S ribosome binds.
- tRNA carries N-formylmethione to first position
- Elongation
- 2 adjacent sites on ribosome: P and A site.A site accepts a new tRNA-AA.Psite holds existing chain peptide transferred from P site tRNA to A-site AA
- enzyme activity is in ribosomal RNA, not protein
- also required: Energy (GTP) and elongation factors
- Termination
- reach a “stop codon” UAG, UAA, or UGA
- no t-RNAs for release, but release factors required
- Net cost: 4 phosphate bonds/amino acid added!
- B. Genetic Code
- AUG = universal “start” codon
- UAG, UAA, UGA = “stop” codons
- A few messages in bacteria use GUG as start, but still need Shine-Dalgarno sequence, stil code for N-formylmethionine.
Classic example: The lac operon (Video)
- gene is regulated by negative control; in absence of specific repressor, gene is transcribed just like constitutive gene. In order to regulate, must add specific block. Must say “no”; otherwise gene is not down-regulated.
- Lactose = milk sugar, disaccharide made of galactose + glucose. In order to metabolize lactose, cells must produce enzyme ß-galactosidase, split lactose into galactose + glucose
- Observation: add lactose to cells: within minutes, ß-galactosidase enzyme appears, also lac permease in membrane, and a third protein, transacetylase. Level of ß-galactosidase enzyme can accumulate to level of 10% of cytoplasmic protein
- Explanation: in absence of lactose (= inducer), lac repressor blocked operator site.
- Lac repressor is allosteric protein. Coded for by another region of DNA (constitutive gene, weak promoter, low level of expression)
- Effector molecule = Inducer is lactose (actually allolactose, or analog such as IPTG) binds to repressor protein, repressor released, RNA is made, all genes turned on as unit
Some genes are regulated by activator proteins
- genes with weak promoters are rarely transcribed.some such genes can be activated by activator protein, causes RNA polymerase to bind more tightly.often there are two components to such regulation: a sensor protein and an activator protein. The activator protein is inactive until phosphorylated by the sensor, then it activates transcription, gene product is made.
Lac operon – in presence of inducer ( lactose)
Lac operon – in absence of lactose
Role of mRNA
Carries coding information for amino acids = codons, 3 adajacent nucleotide bases Example: AAA, AGU, etc.leader sequence on mRNA (called Shine-Dalgarno sequence) binds to complementar sequence on small ribosome subunit.
Role of ribosome
acts as a “decoding box” or “tape player” for the information in mRNA30S & 50S subunits (= 70S)30S has 16S RNA + 21 proteins.50S has 23S & 5S RNA + 34 proteins.
Role of tRNA
- structure: 4 loops, anticodon, AA binding site
- ~ 60 types in bacteria (>100 in mammals)
- only 73-93 nucleotides long
- some modified bases: pseudouridine, inosine, others
- modified after transcription
- extensive hairpin loops
- anticodon site: recognizes codon on mRNA
- AA added by enzyme: AA-tRNA activating enzymes
- ATP required, forms AA-AMP + PP, then AA-tRNA + AMP
| Download this lecture as PDF here |
Transfer of Genetic Material in Bacteria
The process of transfer of genetic material and recombination is very interesting bascterial recombination is given (PPT. an overview of bacterial recombination).The three main mechanisms by which bacteria acquire new DNA are transformation, conjugation, and transduction. Transformation involves acquisition of DNA from the environment, conjugation involves acquisition of DNA directly from another bacterium, and transduction involves acquisition of bacterial DNA via a bacteriophage intermediate.
Transformation
Transformation is the process by which bacteria pick up DNA from their environment. The DNA may come from a variety of sources, but most likely it is the remnants of DNA from dead bacterial cells. 
In order to become successfully transformed, bacteria must be competent. This means that the bacteria are expressing the appropriate enzymes (the ‘transformation machinery’) required to transport the exogenous DNA into the cell. Therefore, the correct genes must be expressed in order to carry out transformation. Expression of these genes depends on the growth conditions: bacteria most likely to be competent are dividing rapidly, but nutrients in the environment are becoming limited. (For more on the control of gene expression, see the module on bacterial gene regulation.
In transformation, a cell surface receptor binds to DNA in the environment. After binding, the DNA is transported across the membrane by the transformation machinery. As this occurs, one strand of the DNA is digested away by an exonuclease, so that the DNA that enters the cell is single stranded. This promotes recombination, as long as the DNA taken up is sufficiently homologous to the host DNA to allow recombination to occur. The recombination that occurs is one-way (non-reciprocal); unlike the exchange of strands diagrammed in the module on recombination, in this case the new DNA will simply replace a strand of the host DNA. The replaced segment of host DNA will be degraded. If the new DNA is of a different allelic nature than the host DNA, a gene conversion event can occur. This is what happened in the example mentioned above: the avirulent strain of S. pneumoniae had a mutation in a gene required for production of the bacterial capsule. Heat killing the virulent cells (which contained the wild-type capsule gene) caused the release of fragments of the dead cells’ genomes. Some of the avirulent cells picked up a piece of DNA containing the wild-type capsule gene, and underwent gene conversion so that they were wild type for that gene, causing them to become virulent.
Conjugation
Conjugation is a mating process involving bacteria. It involves transfer of genetic information from one bacterial cell to another, and requires physical contact between the two bacteria involved. The contact between the cells is via a protein tube called an F or sex pilus, which is also the conduit for the transfer of the genetic material.
Basic conjugation involves two strains of bacteria: F+ and F-. The difference between these two strains is the presence of a Fertility factor (or F factor) in the F+ cells. The F factor is an episome that contains 19 genes and confers the ability to conjugate upon its host cell. Genetic transfer in conjugation is from an F+ cell to an F- cell, and the genetic material transferred is the F factor itself. Here is an overview of the process:
| Basic conjugation occurs between an F+ cell and an F- cell. The difference between these two types of cells is the presence or absence of the F (fertility) factor, which is a circular DNA molecule independent of the bacterial chromosome (the larger circular molecule. |  |
| The F+ cell initiates conjugation by extending an F pilus toward the F- cell. Among the genes present on the F factor are the genes encoding the proteins required for pilus construction. |  |
| The F pilus, when finished, temporarily connects the two cells. On strand of the F factor is nicked, and begins unwinding from the other strand. The nicked strand begins to transfer through the F pilus to the F- cell. As it does so, this strand begins to be replicated, as does circular strand remaining behind in the F+ cell. |  |
| Eventually, the nicked strand completely passes through to the recipient cell, and is completely replicated. This process produces a new F factor in the recipient cell. The pilus is broken, severing the connection between the two cells. Since both cells now contain an F factor, both cells are F+. The new F+ cell (which was the F- cell, can now initiate conjugation with another F- cell. |   |
Recombination rarely occurs with this kind of conjugation. This is because the F factor is not homologous to the DNA in the bacterial chromosome. As we will see, however, there are variations of this basic conjugation process that allow recombination to occur.
Conjugation Involving Hfr Bacteria
Occasionally, the F factor integrates into a random position in the bacterial chromosome. When this happens, the bacterial cell is called Hfr instead of F+. Hfr bacteria are still able to initiate conjugation with F- cells, but the outcome is completely different from conjugation involving F+ bacteria:
| As mentioned above, Hfr cells are formed when the F factor integrates into the bacterial chromosome. This integration occurs at a random location. |
| The Hfr cell is still able to initiate conjugation with an F- cell. |
| When DNA transfer begins, the Hfr cell tries to transfer the entire bacterial chromosome to the F- cell. The first DNA to be transferred is chromosomal DNA, and the last DNA to be transferred will be the F factor DNA. |
| Transfer of the bacterial chromosome is almost never complete. Pili are fairly fragile structures, and shear forces tend to break the pilus, disrupting DNA transfer before the entire chromosome can be transferred. As a result, the F factor itself is almost never transferred to the recipient cell. This cell will remain F-. This cell will receive new DNA from the Hfr cell however, and this new DNA can undergo recombination at a high frequency with the host chromosome, because the DNA sequences will be homologous. In fact, Hfr is short for ‘high frequency recombination’. This recombination can result in gene conversion events, if the transferred DNA and the corresponding region of host DNA contain different alleles of the same gene. |
Mapping Genes on Bacterial Chromosomes
Bacteria, since they are usually haploid, cannot have their chromosomes mapped by the same techniques as eukaryotes (For a reminder of how this works, see the module on linkage and mapping). They can, however, be mapped by using Hfr bacterial conjugation. For example, imagine that an F- cell has mutant alleles of two genes, a and b (the F- would therefore be a-, b-). If this cell undergoes conjugation with an Hfr cell that is a+, b+ (in other words, wild type), the F- cell should undergo gene conversion to a+, b+ when both of those genes have been transferred by conjugation. By determining how long it takes the b gene to transfer after the a gene has transferred, it is possible to get a relative idea of how far apart the two genes are on a chromosome.
The experiment would be done this way: a+, b+ Hfr cells would be mixed with a-,b– F- cells. The time of mixing would be designated ‘time zero’. At regular intervals, a small amount of the mixture would be removed and conjugation would be disrupted using a blender (the shear force of the blender would cause any pili to break). These bacteria would then be tested for gene conversion (for example, if the mutations rendered the F- bacteria auxotrophic, the bacteria could be tested by growing them on minimal medium, or minimal medium supplemented with the necessary nutrient required because of one or the other mutation). If the a gene was converted to wild type at 8 minutes after time zero, and the b gene was converted to wild type at 19 minutes after time zero, then the distance between the two genes would be ’11 minutes’ (because that was the difference in time required to transfer the b gene compared to the a gene). Bacterial map distances are always expressed in minutes, because of this technique.
F’ Conjugation
Just as F factors can occasionally integrate into the bacterial chromosome (producing an Hfr cell from an F+ cell), integrated F factors can occasionally excise themselves from the bacterial chromosome. If this excision occurs properly, the Hfr cell becomes an F+ again. The excision is sometimes sloppy, however, and the F factor takes a small segment of the bacterial chromosome with it. Some of the chromosomal DNA has therefore become associated with the episome. When this happens, the cell is called an F’.
Conjugation involving F’ cells allows for the possibility of recombination, as shown below:
| The F’ cell has a full complement of chromosomal genes; however, some of those genes are now on the episome. F’ cells are able to initiate conjugation with F- cells because of the presence of the F factor. |
| When the F factor begins to transfer its DNA to the recipient cell, it will transfer the small segment of chromosomal DNA as well. |
| Just as in the F+/F- mating, both cells wind up with a copy of the episome. The cell that was F- now has the F factor (along with the piece of chromosomal DNA) and is therefore now F’. This cell, however, also has a complete chromosome, so it will be diploid for the segment of chromosomal DNA on the episome. Such a partially diploid bacterial cell is called a merozygote. The chromosomal DNA on the episome can undergo recombination at high frequency with its homologous sequence on the chromosome. |
Transduction
Transduction involves the exchange of DNA between bacteria using bacterial viruses (bacteriophage) as an intermediate. There are two types of transduction, generalized transduction and specialized transduction, which differ in their mechanism and in the DNA that gets transferred. Before we can address these processes, however, we need to understand the life cycle of a bacteriophage.
When a phage infects a bacterial cell, it injects its DNA into the cell. The viral DNA is replicated numerous times, and viral genes are expressed, producing the proteins that make up the viral capsid (or protein coat) and nucleases that digest the host genome into fragments. The newly replicated viral DNA molecules are packaged into viral capsids, and the bacterial cell is lysed (burst, and therefore killed), releasing hundreds of viral progeny, which then go on to infect other cells.
Generalized Transduction
Sometimes, during bacteriophage replication, a mistake is made, and a fragment of the host DNA gets packaged into a viral capsid. The resulting phage would be able to infect another cell, but it would not have any viral genes, so it would not be able to replicate. The cell infected by this phage would survive, and would have an extra piece of bacterial DNA present, which could undergo recombination with the host chromosome, and perhaps cause a gene conversion event. Because it is a random fragment that gets packaged into the viral capsid, any segment of the bacterial DNA can be transferred this way (hence the name ‘generalized’).
Specialized Transduction
Specialized transduction occurs only with certain types of bacteriophage, such as phage lambda. Lambda has the ability to establish what is called a lysogenic infection in a bacterial cell. In a lysogenic infection, the viral DNA becomes incorporated into the host chromosome, much as the F factor did in Hfr cells. In a lysogenic infection by lambda, the DNA integrates into a very specific spot in the host chromosome. The integrated viral DNA can remain integrated for long periods of time, without disturbing the cell. Under the appropriate conditions (the regulation of this is very complex, so don’t worry about it), the viral DNA will excise itself from the chromosome, and enter the lytic phase, in which the virus replicates just as described above. The cell gets lysed, and new bacteriophage particles are released to infect other cells. As with excision of the F factor (when Hfr cells become F’), sometimes the excision of lambda is sloppy, and some bacteria DNA is excised along with it. When the resulting virus infects another cell, it will pass that bacterial DNA into the cell, along with its own DNA. If the infected cell survives (it can happen; there are bacterial defenses against viral infection), it will contain a new piece of bacterial DNA, which can undergo recombination and possibly cause gene conversion. Because the viral DNA integrates into a specific location, when it excises, the bacterial DNA removed with it will be the same in all cases. Therefore, the DNA transferred to the second cell will be the same segment of the bacterial chromosome. This is why this process is called ‘specialized’ transduction.

Bacterial Recombination: Summary
Bacteria can pick up loose DNA in their environment through the process of transformation. The newly acquired DNA is rendered single stranded, and can recombine with the host chromosome.
- Bacteria can exchange DNA through the process of conjugation. The F factor confers the ability to initiate conjugation. If the F factor alone is transferred, no recombination will occur. Under certain circumstances, chromosomal DNA can be transferred to the recipient cell. In these cases, recombination will occur.
- Bacteria can receive bacterial DNA from viruses through the process of transduction. Bacterial viruses can accidentally pick up pieces of bacterial DNA. When they subsequently infect a cell, they transfer the pice of bacterial DNA, which can undergo recombination with the host bacterial chromosome.
- The result of recombination in the above cases may be gene conversion, in which a mutant allele becomes wild-type or vice versa.
- Conjugation involving Hfr bacteria can be used to map genes along the bacterial chromosome. This done by determining in what order genes are transferred during conjugation, waht the time difference is between the transfer of genes.
Bacteria do not reproduce sexually but can acquire new DNA through transformation, transduction or conjugation.These natural processes have been modified so that DNA can be deliberately incorporated into host microbes- even genes that would normally never be transferred this way.
| Download this lecture as PDF here |
Genetic Engineering: Genetic engineering is a laboratory technique used by scientists to change the DNA of living organisms.
DNA is the blueprint for the individuality of an organism. The organism relies upon the information stored in its DNA for the management of every biochemical process. The life, growth and unique features of the organism depend on its DNA. The segments of DNA which have been associated with specific features or functions of an organism are called genes.
Molecular biologists have discovered many enzymes which change the structure of DNA in living organisms. Some of these enzymes can cut and join strands of DNA. Using such enzymes, scientists learned to cut specific genes from DNA and to build customized DNA using these genes. They also learned about vectors, strands of DNA such as viruses, which can infect a cell and insert themselves into its DNA.
With this knowledge, scientists started to build vectors which incorporated genes of their choosing and used the new vectors to insert these genes into the DNA of living organisms. Genetic engineers believe they can improve the foods we eat by doing this. For example, tomatoes are sensitive to frost. This shortens their growing season. Fish, on the other hand, survive in very cold water. Scientists identified a particular gene which enables a flounder to resist cold and used the technology of genetic engineering to insert this ‘anti-freeze’ gene into a tomato. This makes it possible to extend the growing season of the tomato.
Plasmids: A plasmid is an extra chromosomal DNA molecule separate from the chromosomal DNA which is capable of replicating independently from the chromosomal DNA. In many cases, it is circular and double-stranded. Plasmids usually occur naturally in bacteria, but are sometimes found in eukaryotic organisms (e.g., the 2-micrometre-ring in Saccharomyces cerevisiae).
Plasmid size varies from 1 to over 1,000 kilobase pairs (kbp). The number of identical plasmids within a single cell can range anywhere from one to even thousands under some circumstances. The number of plasmids in a cell generally remains constant from generation to generation.
Properties of Plasmids
• Circular DNA elements, always double-stranded DNA, Supercoiled
• Can occur in as few as 1 copy per cell (single copy plasmids) to as many as several dozen (multicopy plasmids).
• Variable sizes; small plasmids about 0.1% size of host chromosome, large plasmids can be as much as 10% the size of host chromosome. Smaller plasmids have few genes (30 or less). Size ranges from 1000 bp (1 kbp) to 1000 kbp.
• Ubiquitous; almost all cells isolated in nature carry plasmids, often more than one kind. (In E. coli alone, more than 300 different plasmids isolated.)
• Have a replicon (origin for DNA replication), number of copies per cell regulated. Large plasmids typically only 1-5 copies/cell (stringent control); small plasmids ~10-50 copies/cell (relaxed control)
• Many plasmids are incompatible; if one is present, cell cannot support another plasmid of same compatibility group.
• Not essential to cell under all circumstances; can be “cured” by agents that impair DNA replication —-> cured cell lacking plasmid. Can be spontaneously lost over time unless some selection makes plasmid valuable to cell.
• Extend range of environments in which a cell can live (e.g., by degrading antibiotics, or providing enzymes for digestion of novel catabolites).
Examples of Plasmid genes
- Antibiotic resistance genes (enzymes that modify or degrade antibiotics) — plasmids with these genes are called R factors
- Heavy metal resistance (enzymes that detoxify metals by redox reactions)
- Growth on unusual substrates (enzymes for hydrocarbon degradation, etc.)
- Restriction/modification enzymes (protect DNA, degrade unprotected DNA)
- Bacteriocins (proteins toxic to other bacteria lacking the same plasmid)
- Toxins (proteins toxic to other organisms; e.g. humans) — called virulence plasmids.
Some Examples:
- Staph aureus virulence factors: coagulase, hemolysin, enterotoxin, others
- pathogenic E. coli strains: hemolysin, enterotoxin
Proteins that mediate plasmid transfer to uninfected strains
There are two categories of plasmids. Stringent plasmids replicate only when the chromosome replicates. This is good if you are working with a protein that is lethal to the cell. Relaxed plasmids replicate on their own. This gives you a higher ratio of plasmids to chromosome. Some of the traits coded by plasmids include:

Classification of Plasmids
1. Transfer properties –
a. Conjugative plasmids – Conjugative plasmids are those that mediated conjugation. These plasmids are usually large and have all the genes necessary for autonomous replication and for transfer of DNA to a recipient (e.g. genes for sex pilus).
b. Nonconjugative plasmids – Nonconjugative plasmids are those that cannot mediate conjugation. They are usually smaller than conjugative plasmids and they lack one or more of the genes needed for transfer of DNA. A nonconjugative plasmid can be transferred by conjugation if the cell also harbors a conjugative plasmid.
2. Phenotypic effects –
a. Fertility plasmid (F factor)
b. Bacteriocinogenic plasmids – These plasmids have genes which code for substances that kill other bacteria. These substances are called bacteriocins or colicins.
c. Resistance plasmids 7 factors) – These plasmids carry antibiotic resistance genes.i) Origin – The origin of the R factors is not known. It is likely that they evolved for other purposes and the advent of the antibiotic age provided a selective advantage for their wide-spread dissemination.
ii) Structure – R plasmids are conjugative plasmids in which the genes for replication and transfer are located on one part of the R factor and the resistance genes are located on another part as illustrated in Figure.
RTF (Resistance Transfer Factor) – carries the transfer genes.
R determinant – carries the resistance genes. The resistance genes are often parts of transposons.
Mode of action of resistance genes –
a) Modification (detoxification) of antibiotic – e.g. β-lactamase
b) Alteration of target site – e.g. Streptomycin resistance
c) Alteration of uptake – Tetracycline resistance
d) Replacement of sensitive pathway – e.g. new folic acid pathway for resistance to sulfa drugs.
Plasmids are easy to manipulate and isolate using bacteria. They can be integrated into mammalian genomes, thereby conferring to mammalian cells whatever genetic functionality they carry. Thus, this gives you the ability to introduce genes into a given organism by using bacteria to amplify the hybrid genes that are created in vitro. This tiny but mighty plasmid molecule is the basis of recombinant DNA technology.
Episome: Episome is a unit of genetic material composed of a series of genes that sometimes has an independent existence in a host cell and at other times is integrated into a chromosome of the cell, replicating itself along with the chromosome. Episomes have been studied in bacteria. One group of episomes are actually viruses that infect bacteria. As autonomous units they destroy host cells, and as segments integrated into a chromosome they multiply in cell division and are transferred to daughter cells. Episomes called sex factors determine whether chromosome material will be transferred from one bacterium to another. Other episomes carry genes that make bacteria resistant to the inhibitory action of antibiotics.
Transposons : Are sequences of DNA that can move or transpose themselves to new positions within the genome of a single cell. The mechanism of transposition can be either “copy and paste” or “cut and paste”. Transposition can create phenotypically significant mutations and alter the cell’s genome size. Barbara McClintock‘s discovery of these jumping genes early in her career earned her a Nobel prize in 1983. Transposons make up a large fraction of the C-value of eukaryotic cells. Transposons are often considered “junk DNA“. In Oxytricha, which has a unique genetic system, they play a critical role its development. Transposons are very useful to researchers as a means to alter DNA inside a living organism.
| Download this lecture as PDF here |
Genetic engineering – was made possible through a series of scientific advances including the discovery of DNA and the creation of the first recombinant bacteria in 1973, i.e., E .coli expressing a salmonella gene. This led to concerns in the scientific community about potential risks from genetic engineering.
A genetically modified organism (GMO) or genetically engineered organism (GEO) is an organism (plant or animal) whose genetic material has been altered using genetic engineering techniques. These modifications are generally used to benefit medicine or food production. There is controversy about these techniques, as there are fears of tampering with an organism’s evolution, and the long-term irreversible effects that could come from that tampering.
The general principle of producing a GMO is to add a lot of genetic material into an organism’s genome to generate new traits. These techniques, generally known as recombinant DNA technology, use DNA molecules from different sources, which are combined into one molecule to create a new set of genes. This DNA is then transferred into an organism, giving it modified or novel genes. Transgenic organisms, a subset of GMOs, are organisms which have inserted DNA that originated in a different species.
Genetically modified bacteria were the first organisms to be modified in the laboratory, due to their simple genetics. These organisms are now used for several purposes, and are particularly important in producing large amounts of pure human proteins for use in medicine.The first example of this occurred in 1978 when Herbert Boyer working at a University of California laboratory took a version of the human insulin gene and inserted into the bacterium Escherichia coli to produce synthetic “human” insulin.The drug industry has made good use of this discovery in its quest to cure diabetes. Similar bacteria have been used to produce clotting factors to treat haemophilia, and human growth hormone to treat various forms of dwarfism. These recombinant proteins are safer than the products they replaced, since the older products were purified from cadavers and could transmit diseases. Indeed the human-derived proteins caused many cases of AIDS and hepatitis C in haemophilliacs and Creutzfeldt-Jakob disease from human growth hormone.
For instance, the bacteria which cause tooth decay are called Streptococcus mutans. These bacteria consume leftover sugars in the mouth, producing lactic acid that corrodes tooth enamel and ultimately causes cavities. Scientists have recently modified Streptococcus mutans to produce no lactic acid. These transgenic bacteria, if properly colonized in a person’s mouth, could reduce the formation of cavities. Transgenic microbes have also been used in recent research to kill or hinder tumors, and to fight Crohn’s disease. Genetically modified bacteria are also used in some soils to facilitate crop growth, and can also produce chemicals which are toxic to crop pests.
Moving Genes between Species—how to do……
- The process by which scientists introduce new genetic material into a microorganism is called molecular or gene cloning or genetic engineering.
- It involves the isolation of DNA from a source other than the microorganism itself. Source organisms span the world of living things, from microbes to plants to animals, including humans. Scientists obtain source DNA in several different ways: by disrupting cells of the target microbe (or plant or animal) and fragmenting it into small pieces, by synthesizing it from an RNA template using an enzyme called reverse transcriptase, or by knowing the specific gene sequence and synthesizing it directly in the laboratory.
- Once obtained, the pieces of DNA are inserted into a small genetic component that has the ability to make copies of itself (replicate) independently from the microbial genome. This self-replicating unit is called a cloning vector. Although these genetic elements exist naturally in the form of plasmids and bacterial viruses, many of the ones used today have been altered to improve their properties for transferring genes. Restriction enzymes, which nick the donor DNA and the cloning vector at specific sites, and DNA ligase, which attaches the donor DNA to the cloning vector, allow the source genes of interest to be inserted into the cloning vector without disrupting its ability to replicate.
- The next step in the process is the introduction of the cloning vector with its segment of new DNA into a living cell. Bacteria have the ability to transport DNA into their cells in a process called transformation, and this ability is commonly exploited to achieve this goal. Getting the DNA into the cell, however, is only the beginning. No transformation is 100 percent efficient, and so the bacteria that receive the gene(s) of interest must be separated from those that did not. One of the best studied and most commonly used cloning vectors, pBR322, is especially useful for this purpose, as it contains several genes for antibiotic resistance. Hence, any cell transformed with DNA containing pBR322 will be antibiotic resistant, and thus can be isolated from similar cells that have not be so transformed by merely growing them in the presence of the appropriate drugs. All that remains is to identify bacteria that are producing the product of the desired gene(s), and cloning is a success.
- The introduction of human genes into bacteria has several complicating wrinkles that make cloning them even more challenging. For example, a bacterial gene codes for a protein from start to finish in one long string of nucleotides, whereas human cells have stretches of noncoding nucleotides called introns within their genes. Bacteria do not have the same ability as human cells to remove these introns when producing proteins from the gene, and if the introns are not removed, the intended protein cannot be produced. This, along with other complications, has been overcome using many of the tools of genetic engineering.
Uses of GMOs: Examples of GMOs are highly diverse, and include transgenic (genetically modified by recombinant DNA methods) animals such as mice, fish, transgenic plants, or various microbes, such as fungi and bacteria. The generation and use of GMOs has many reasons, chief among them are their use in research that addresses fundamental or applied questions in biology or medicine, for the production of pharmaceuticals and industrial enzymes, and for direct, and often controversial, applications aimed at improving human health (e.g., gene therapy) or agriculture (e.g., golden rice). The term “genetically modified organism” does not always imply, but can include, targeted insertions of genes from one into another species. For example, a gene from a jellyfish, encoding a fluorescent protein called GFP, can be physically linked and thus co-expressed with mammalian genes to identify the location of the protein encoded by the GFP-tagged gene in the mammalian cell. These and other methods are useful and indispensable tools for biologists in many areas of research, including those that study the mechanisms of human and other diseases or fundamental biological processes in eukaryotic or prokaryotic cells.
Transgenic microbes:
Bacteria were the first organisms to be modified in the laboratory, due to their simple genetics. These organisms are now used in a variety of tasks, and are particularly important in producing large amounts of pure human proteins for use in medicine.
Bacteria-synthesized transgenic products
- Insulin
- Interferon
- Hepatitis B vaccine
- Tissue plasminogen activator
- Human growth hormone
- Ice-minus bacteria
Did you know that this is a is a nickname given to a variant of the common bacterium Pseudomonas syringae (P. syringae). This strain of P. syringae lacks the ability to produce a certain surface protein, usually found on wild-type P. syringae. The “ice-plus” protein (In a protein, “Ice nucleation-active” protein) found on the outer bacterial cell wall acts as the nucleating centers for ice crystals. This facilitates ice formation, hence the designation “ice-plus.” The ice-minus variant of P. syringae is a mutant, lacking the gene responsible for ice-nucleating surface protein production. This lack of surface protein provides a less favorable environment for ice formation. Both strains of P. syringae occur naturally, but recombinant DNA technology has allowed for the synthetic removal or alteration of specific genes, enabling the creation of the ice-minus strain. Modifying P. syringae may have unexpected consequences for climate. A study has shown that its ice nucleating proteins may play an important part in causing ice crystals to form in clouds. If humans increase the frequency of bacteria lacking these proteins then it may affect rainfall
Commercial Applications
- Transgenic microbes have many commercial and practical applications, including the production of mammalian products. A company called Genentech was among the earliest and most successful commercial enterprises to use genetically engineered bacteria to produce human proteins. Their first product was human insulin produced by genetically engineered Escherichia coli. A variety of other human hormones, blood proteins, and immune modulators are now produced in a similar fashion, in addition to vaccines for such infectious agents as hepatitis B virus and measles.
- Another promising application of genetically engineered microbes is in environmental cleanup, or biomediation. Scientists have discovered many naturally occurring genes that code for enzymes that degrade toxic wastes and wastewater pollutants in bacteria. Examples include genes for degrading chlorinated pesticides, chlorobenzenes, naphthalene, toluene, anilines, and various hydrocarbons. Researchers are using molecular cloning to introduce these genes from several different microbes into a single microbe, creating “super microbes” with the ability to degrade multiple contaminants.
- Ananda Chakrabarty created one of the first microbes of this nature in the early 1970s. He introduced genes from several different bacteria into a strain of Burkholderia cepacia, giving it the ability to degrade toxic compounds found in petroleum. This microbe offered a potential alternative to skimming and absorbing spilled oil. Chakrabarty’s genetically modified bacterium has never been used, however, due to public concerns about the release of genetically engineered microbes into the environment. The microbe did, on the other hand, play an important role in establishing the biotechnology industry. The U.S. Patent Office granted Chakrabarty the first patent ever for the construction and use of a genetically engineered bacterium. This established a precedent allowing biotechnology companies to protect their “inventions” in the same way chemical and pharmaceutical companies have done in the past.
In addition to bacteria being used for producing proteins, genetically modified viruses allow gene therapy. Gene therapy is a relatively new idea in medicine. A virus reproduces by injecting its own genetic material into an existing cell. That cell then follows the instructions in this genetic material and produces more viruses. In medicine this process is adapted to deliver a gene that could cure disease into human cells. Although gene therapy is still relatively new, it has had some successes. It has been used to treat genetic disorders such as severe combined immunodeficiency, and treatments are being developed for a range of other incurable diseases, such as cystic fibrosis, sickle cell anemia, and muscular dystrophy.
Genetically modified bacteria are also used in agriculture to facilitate crop growth, and can also produce chemicals which are toxic to crop pests.
Transgenic animals
Transgenic animals are used as experimental models to perform phenotypic tests with genes whose function is unknown or to generate animals that are susceptible to certain compounds or stresses for testing in biomedical research. Other applications include the production of human hormones, such as insulin. Frequently used in genetic research are transgenic fruit flies (Drosophila melanogaster) as genetic models to study the effects of genetic changes on development. Transgenic mice are often used to study cellular and tissue-specific responses to disease.
Transgenic plants
Transgenic plants have been developed for various purposes. Most of transgenic plants were created for research purposes and were not intended for eventual commercialization. From these few which have reached the market the most common transgenic traits include 1) resistance to pests or herbicides, 2) improved product shelflife. In the near future crops with improved nutritional value and with resitance to harsh environmental conditions might reach the marketplace. Since the first commercial cultivation of GM plants in 1996, GM plants tolerant to the herbicides glufosinate or glyphosate, and producing the Bt toxin, an insecticide, have dominated the agriculutral seed market for corn and other crops (soybean, cotton). Recently, a new generation of GM plants promising benefits for consumers and industry purposes is entering the market. Whenever GM plants are grown on open fields without containment there are risks that the modification will escape into the general environment. Most countries require biosafety studies prior to the approval of a new GM plant release, usually followed by a monitoring programme to detect environmental impacts. Especially in Europe, the coexistence of GM plants with conventional and organic crops has raised many concerns. Since there is separate legislation for GM crops and a high demand from consumers for the freedom of choice between GM and non-GM foods, measures are required to separate foods and feed produced from GMO plants from conventional and organic foods. European research programmes such as Co-Extra, Transcontainer and SIGMEA are investigating appropriate tools and rules.
The use of GMOs has sparked significant controversy in many areas. Some groups or individuals see the generation and use of GMO as intolerable meddling with biological states or processes that have naturally evolved over long periods of time (although many crops and animals have been modified by humans via unnatural selection over the last several thousand years), while others are concerned about the limitations of modern science to fully comprehend all of the potential negative ramifications of genetic manipulation.
While some groups advocate the complete prohibition of GMOs, others call for mandatory labeling of genetically modified food or other products. Other controversies include the definition of patent and property pertaining to products of genetic engineering and the possibility of unforeseen local and global effects as a result of transgenic organisms proliferating. The basic ethical issues involved in genetic research are discussed in the article on genetic engineering.
| Download this lecture as PDF here |
The field of soil microbiology was explored during the very last part of 19th century. The establishment of the principal roles that microorganisms play in the biologically important cycles of matter on earth: the cycles of nitrogen, sulphur and carbon was largely the work of two men, S. Winogradsky (1856-1953) and M.W. Beijerinck (1851–1931). S. Winogradsky, Russian and regarded by many as the founder of soil microbiology, discovered nitryfiying bacteria (1890-91); described the microbial oxidation of H2S and sulphur (1887); developed the contributed to the studies of reduction of nitrate and symbiotic nitrogen fixation; and, originated the nutritional classification of soil microorganisms into autochtonous (humus utilizers) and zymogenous (opportunistic) groups.
Almost equally important was the work of M.W. Beijerinck, a Hollander, who isolated the agents of symbiotic (1888) and non-symbiotic aerobic (1901) nitrogen fixation. However, the greatest contribution of Beijerinck was a new and profoundly important technique: enrichment culture technique: to isolate and study various physiological types of various microorganisms from natural samples through the use of specific culture media and incubation conditions.
Bacteria– more dominant group of microorganisms in the soil and equal to one half of the microbial biomass in soil. Majority are Heterotrophs. (Common soil bacteria – Arthrobacter, Bacillus, Clostridium, Micrococcus).




ROD SHAPED BACTERIA
 SPHERICAL BACTERIA
SPHERICAL BACTERIA




Actinomycetes – intermediate group between bacteria and fungi. Numerous and widely distributed in soil. Abundance is next to bacteria. 104 – 108/g soil. 70% of soil actinomycetes are Streptomyces. Many of them are known to produce antibiotics. Population increases with depth of soil.
Actinomycetes – intermediate group between bacteria and fungi. Numerous and widely distributed in soil. Abundance is next to bacteria. 104 – 108/g soil. 70% of soil actinomycetes are Streptomyces. Many of them are known to produce antibiotics. Population increases with depth of soil.
Fungi: More numerous in surface layers of well-aerated and cultivated soils-dominant in acid soils. Common genera in soil are Aspergillus, Mucor, Penicillium Trichoderma, Alternaria, Rhizopus.




Algae – found in most of the soils in number ranges from 100 to 10,000 per g.


Protozoa: Unicellular – population ranges from 10,000 to 100,000 per g of soil. Most of the soil forms are flagellates, amoebae or ciliates. Derive their nutrition by devouring soil bacteria. Abundant in upper larger of the soil. They are regulating the biological equilibrium in soil.



Importance
- Involved in nutrient transformation process
- Decomposition of resistant components of plant and animal tissue
- Role in microbial antagonism
- Participate in humus formation
- Predator of nematodes
- Surface blooming reduces erosion losses
- Improve soil structure
- Involved soil structure
- Maintenance of biological equilibrium
actors influencing activities of soil microorganisms: Soil microorganisms are influenced by various factors. Chief factors are,fertility levelSoil moistureSoil airsoil temperatureOrganic matterH ion concentrationCultural factors.
| Download this lecture as PDF here |
The term soil generally refers to the loose material of the earth surface and is the region that supports the plant life. It consists of five major components such as mineral matter, water, air, organic matter and living organisms. The proportion of these components varies with soil type and other soil conditions. To maintain the level of these components it is essential that they undergo a regular process of recycling. This process of recycling through various transformations is brought about by different microorganism.
A- PS
B- Respiration / plant
C- Respiration / Animals
D- Autotrophs
E- Respiration / Microbial mineralization

The most important element in the biological realm and substance that serve as the cornerstone of the cell structure is carbon. It constituents about 40-50% of all living organisms, yet the ultimate source is the CO2 that exist in a perennially short supply, only 0.03% of the earth’s atmosphere, which undergo a cyclic change from an oxidized to reduced state.
Carbon (CO2) is constantly (reduced into organic carbon compounds) being fixed into organic form by photosynthetic organisms (photosynthesis). Once bound, the carbon becomes unavailable for use in generation of new plant life. It is thus essential for the carbonaceous materials to be decomposed and returned to the atmosphere. It is estimated that 1.3×1014 kg CO2 is fixed annually in the biosphere. To the lesser extent CO2 is also fixed through the agency of photosynthetic bacteria and other chemolithotrophs with the convertion of so much of the plant available carbon to organic form each year and the limited supply in the air, it is apparent that the major plant nutrient element would become exhausted in the absence of microbial transformation.
The carbon cycle revolves about CO2 and its fixation and regeneration. The green plants utilize CO2 as their sole carbon source, and the carbonaceous matter synthesized serves to supply carbon to other heterotrophic organisms and animals. Upon the death of plants and animals, microbes assume a dominant role in carbon cycle. The dead tissues are degraded and transformed into microbial cells and humus or soil organic fraction. Further decomposition of these materials leads to the production of CO2 and once again it is recycled.
Organic matter decomposition (Aerobic decay)
Soil organic matter
The organic matter subjected to microbial decay in soil comes from several sources like plant remains, forest litter, incorporation of plant and animal tissues and excretory products. The chemistry of organic matter is clearly very complex, and investigations of the transformations and the responsible organisms have therefore been extremely interesting. The organic constituents of the plants are commonly divided into six categories.
a) Cellulose – Most abundant 15-60% of the dry weight
b) Hemicellulose – 10-30% of the plant dry weight
c) Lignin – 5 – 30 % of the plant dry weight
d) Water soluble fraction – 5-30%, included simple sugar, a. acids,
e) Ether and alcohol soluble constituents, a fraction containing fat, oils, waxes, resins and a number of pigments
f) Proteins.
As the plant ages, the content of water soluble constituents, proteins and minerals decreases and the % of abundance of cellulose, hemicellulose and lignin rises.
Soil organic matter comprises residues of plant and animals and these compounds occur in soil in close combination with inorganic substances. Animals and plant residues are made up of complex carbohydrates, simple sugars, starch, cellulose, hemicellulose, pectins, gums, mucilage, proteins fats, oils, waxes, resins, alcohols, aldehydes , ketones, organic acids, lignin, phenols, tannins, hydrocarbons, alkaloids, pigments etc.
- The soil microorganism play important role in the decomposition of soil organic matter.
- Bacteria are the dominant group – mostly heterotrophic organisms (use energy from organic sources such as sugars, starch, cellulose and protein) – are involved. Autotrophic organism which occupy a small portion of the biomass in soil (and use inorganic sources such as Fe and S) are not directly involved in organic matter decomposition.
- Actinomycetes grow on complex substances such as keratin, chitin and other complex polysaccharides and play active role in humus formation.
- Soil fungi are mostly heterotrophos and use organic residues easily
- Soil algae contribute a small amount of organic matter through their biomass, but they do not have any active role in organic matter decomposition.
Organic matter decomposition serves two important functions
a) Provide energy for growth
b) Supply carbon for the formation of new cell materials
Hence only heterotrophs are actively involved in the process of decomposition. The relationship between organic matter and plant growth may be direct or indirect.
- Organic matter is a natural substrate for saprophytic micro organism and provides nutrition to plants indirectly through the activity of soil microorganisms
- It is essential for the formation of soil aggregates and hence soil structure which ultimately determines the soil aeration and rooting habit of plants
- Organic matter helps in the conservation of soil nutrients by preventing erosion and surface run off of nutrients.
Carbon assimilation
The process of converting substrate to protoplasmic carbon is known as assimilation. Under aerobic conditions 20-40% of the substrate carbon is assimilated, the remainder is released as CO2. Fungi are more efficient, in their metabolism, since they convert carbon into cell carbon as filaments and release less of CO2. 30-40% of which is used to form new mycelium during the decomposition. Compared to fungi, bacteira are less efficient. Aerobic bacteria are less efficient than anerobic bacteria.
C. Mineralization
- Conversion of organic Carbon substance to inorganic form of carbon.
Immobilization
Assimilation of nutrients and is the mechanism by which micro organism reduce the quantity of plant available nutrient in soil. Mineralization is considered well than immobilization.
During the decomposition of organic matter three separate simultaneous processes can be distinguished. The important changes during decomposition are:
- Plant and animal tissues constituents disappear under the influence of enzymes
- Synthesis of new microbial cells so that proteins, polysaccharides and nucleic acids typical of bacteria and fungi appear.
- Third, certain end products of the breakdown are excreted into surroundings there to accumulate or to be further metabolized.
Importance of organic matter decomposition
- Important function is the breakdown of organic matter by which CO2 available for photosynthesis is replenished
- Any compound that is synthesized biologically is subject to destruction by soil inhabitants, otherwise the compounds would have accumulated in vast amounts on the earth’s surface
- Since, organic matter degradation is a property of all heterotrophs, it is commonly used to indicate the level of microbial activity.
Methods to evaluate the decomposition rate
- Measurement of CO2 evolution or O2 uptake
- Determination of decrease in organic matter either chemically or by weight loss
- Observations on disappearance of specific constituents such as cellulose, hemicellulose or lignin.
Changes during organic matter decomposition
As a result of development of mixed flora on chemically complex natural products, some components quickly disappear while others are less susceptible to microbial enzymes and persist. The water soluble fraction contains the least resistant plant components and is thus the first to be metabolized. Cellulose and hemicellulose on the other hand disappear not as quickly as water soluble substances, but their persistence usually is not too great. The lignins are highly resistant and consequently become relatively more abundant in the residual, decaying organic matter.
- At aerobic conditions when carbonaceous substrates are incorporated into soil, immediate drop in O2 and an increase in CO2 content of soil air occurs.
- Change in (O) (H) oxidation reduction potential (En) – it is shifted to a more reduced condition (fall in oxidation reduction potential).
The end products of decomposition are – CO2, H2O, NO3, SO4, CH4, NH4 and H2S depending on the availability of air.
Factors influencing the organic matter decomposition
- Organic matter level of the soil
- Cultivation
- Temperature
- Moisture
- pH
- Depth
- Aeration
- Nature and abundance of micro organic involved.
- The extent of availability of C, N, P and K presence of inhibitory substance.
C: N ratio
- Nitrogen is a key nutrient substance for microbial growth
- If N content of the substrate is high it is readily utilized and decomposition is faster
- If N is poor decomposition is slower, needs additional N
- Protein rich substrates are readily decomposed
- Low N or wide C:N ratio results slow decay
- Optimum level of C: ratio for maximum decomposition is 20-25(1.4-1.7% N)
- Less than this range, more microbial cells, faster mineralization and it likely exceeds immobilization
- Wider the ratio, lesser microbial cells, slower the immobilization and mineralization increases gradually, resulting in accumulation of Ammonia and Nitrates
- Microbes scavenge the soil solution to obtain enough N
- At optimum level, there must be an equilibrium between Mineralization and Immobilization
- Soil N level constrains the maintenance of C:N (organic carbon /soil o.m)
- To make sound soil management
- Arable surface -10:1
- Sub soil -lower
Anaerobic decay / decomposition
The main products of aerobic carbon mineralization are CO2, water, cells and humus components. In the absence of O2 organic carbon is incompletely metabolized, intermediary substances accumulate, and abundant quantities of CH4 and smaller amounts of H2 are evolved. Energy yield during anaerobic fermentation is low, resulting in fewer microbial cells per unit of organic carbon degraded. Consequently, organic matter breakdown is consistently slower under total anaerobiosis than in environments containing adequate O2. The rate in water logged soils is intermediate between the two extremes.
When a soil is water logged or flooded there is a shift from aerobic to anaerobic transformation. Formation and accumulation of organic acids viz., acetic, formic, butyric, lactic and succinic acids appear too, these are frequently detrimental to root development. Organic acids accumulate because of the fermentative character of the microflora of wet soils. The an aerobic carbon transformation are thus characterized by the formation of organic acids, alcohols, CH4 and CO2 as major end products.
Under anaerobic conds, decomposition of organic residues takes place by the activity of both mesophilic, thermophilic microorganism resulting in the production of CO2, H, ethyl alcohol, and organic acids. Among mesoophilis, bacteria are more active than fungi or actinomycetes in cellulolytic activity. They belong to genus Clostridium which are numerous in manure pits but rarely encountered in cultivated arable soils. In compost pits both meso and thermopholic (bacterial and actinomycetes) are important in the breakdown of cellulose substances.
The primary microbial colonisers initially break down the complex CH2O and proteins into organic acids and alcohols. At a later stage, the methane bacteria which are strict anaerobes begin to act upon the secondary substrate chiefly lactic and butyric acids and ferment them into CH4 and CO2.
Humus
- A dark coloured and fairly stable soil organic matter with known and unknown physical and chemical properties
- It is an integral part of the organic matter complex in soil
- Humus can be defined as lingo protein complex containing approximately
45 % – lignin compounds
35% – amino acids
11% – carbohydrates
4% – cellulose
7% – Hemicellulose
3% – fats, wax, resins
6% – other miscellaneous substances, including plant growth substances and inhibitors.
- Age and composition of the humus are dependent on its origin and environment.
- Bacterial and algal protoplasm contribute a good deal to the nutritive value of humus
- Soil micro organism take part in humus formation. Some fungi such as Penicillium, Aspergillus and actinomycetes produce dark humus like substances which serve as structural units for the synthesis of humic substances.
Benefits of humic substances
- Improved seed germination, root growth, uptake of minerals by plants and other physiological effects on plant growth
- Increases the enzyme activities involved in plant metabolism. Since humic acid serves as hydrogen acceptors.
- Increases the cytochrome oxidase activity in root systems results in growth stimulatory effect (on roots)
- Chelating effect – on trace elements Fe uptake by roots
- Vigour and yield of plants enhanced
- Humic acid known to influence the grown and proliferation of micro organism
- Aspergillus niger, Peni, Bacillus sp., Azotobacter are enhanced
The organic fraction of soil, often termed humus. It is a product of synthetic and decomposing activities of the microflora. Since it contains the organic C and N needed for microbial development, it is the dominant food reservoir. Because humus is both a product of microbial metabolism and an important food source, the organic fraction is of special interest.
Humus formation
- Once the plant or animal remains fall on or are incorporated into the soil, they are subjected to decomposition
- From the original residues, a variety of products are formed
- As the original materials and the initial products undergo further decomposition, they are converted to brown or black organic complexes
- At this stage any trace of the original remains no longer remains
- The native organic fraction originates from two sources: the original plant debris entering the soil and the micro organism with in the soil body. The micro organism in soil body, work upon the former and synthesize microbial protoplasm and new compounds that become part of the organic fraction.
- Humus exist in a dynamic state
- Chemistry of humus is complex
- It has been pointed out that the organic fraction is derived from
- Plant constituents that are modified by the microflora.
- Constituents of microbial cells and products of microbial metabolism are relatively resistant to decay and therefore persist for sometime after death of organism.
Interms of specific elements
The organic fraction contains compounds of C, H, O, N, P, S and small amounts of other elements. Only a small portion of the total is soluble in water, but much can be brought into solution by alkali.
Interms of type of compounds
Humus contains a number of polymerized substances, aromatic, molecules, polysaccharides, ascorbic acids, polymers of uronic acids and P containing compounds.
No definite composition can be assigned. It should be considered as a portion of the soil that is composed of a heterogenous group of substances, most having an unknown parentage and an unknown chemical structure.
Lignin and lignin derived molecules have long been considered to be of significance in the formation of humus.
It is possible either that simple aromatics released in the microbial attack on lignin polymerize to yield constituents of the soil organic fraction or those partially altered lignins itself give rise to humus constituents. The monomeric portions of humus are similar to the constituents of lignin.
Degradation processes
(1) Cellulose is a CH2O composed of glucose units bound by β-linkage at carbon 1 and 4 of the sugar molecule. The cellulose concentration of higher plants is never fixed and the concentration. It is a polymer of glucose and is might abundant organic material in nature changes with age and type of plants. Woody materials have more cellulose and succulent tissues had poor, but the concentration increases as the plant matures. Cellulose breakdown in soil is influenced by several environmental factors.
Aerobic organism converts cellulose to 2 major products: CO2 + cell substance, but certain group releases small amount of organic acids. It is however resistant to microbial decomposition. When cellulose is associated with pentosans (xylan, mannans) it undergoes rapid decomposition. When associated with lignin, the decomposition rate is very low. Degradation is by the enzymes that converts cellulose into glucose. (Exoenzymes)
Exoglucanase
Endoglucanase
β – glucosidase (cellulse complex)
Exo glucanase![]()
![]() Native cellulose Amorphos cellulose + cellobiose
Native cellulose Amorphos cellulose + cellobiose![]() Endoglucanase Endogluconase
Endoglucanase Endogluconase
β-glucosidase ![]() D- Glucose Cellobiose
D- Glucose Cellobiose
(cellobiase)
Most cellulolytic bacteria do not excrete significant amounts of cellulase but fungi are found to excrete these enzymes. The soluble sugars released by enzymatic hydrolysis are later utilized by the same or other micro organism for biosynthetic purpose.
(2) Hemicellulose
It is a polymer of simple sugars such as pentose, hexose and uronic acids. They may be either homo or hetero polymers. When they are added to soil, degradation takes place at faster rate in initial stages. The hemicelluloses such as mannans are decomposed rapidly while galactons (polymer of galactose) are decomposed slowly. Many soil micro organisms utilize hemicellulose in both aerobic as well as anaerobic conditions. The microbial degradation occurs through the agency of extracellular enzymes called hemicellulases.
(3) Lignin
Lignin is the third most abundant costituent of plants. It consists of heterocyclic aromatic organic molecules containing C, H and O. The degradation is very slow and rate of decomposition depends on the presence of other compounds such as cellulose and hemicellulose acid. Lignin is highly resistant to microbial degradation. Degradation is a complex process.
- Lignin –à coniferyl ether –à coniferyl alcohol –à coniferyl aldehyde –à vanillin –à vannillic acid –à protocatechuic acid –à ring cleavage
Genera of microorganisms capable of utilizing different components of organic mater as reported by several workers: F-Fungi; B-Bacteria; A-Actinomycetes
| Nature of substrate in organic matter |
| Genera of microorganisms |
Cellulose | F | Alternaria, Aspergillus, Chaetomium, Coprinus, Fomes, Fusarium, Myrothecium, Penicillum, Polyporus, Rhizoctonia, Rhizopus, Trametes, Trichoderma, Trichothecium, Verticillium, Zygorynchus |
B | Achrombacter, Angiococcus, Bacillus, Cellfalcicula, Cellulomonas, Cellvibrio, Clostrium, Cytophaga, Polyangium, Pseudomonas, Sorangium, Sporocytophaga, Vibrio | |
A | Micromonospora, Nocardia, Streptomyces, Streptosporangium | |
Hemicellulose | F | Alternaria, Fusarium, Trichothecium, Aspergillus, Rhizopous, Zygorynchus, Chateomium, Helminthosporium, Penicillium, Coriolus, Fomes, Polyporus |
B | Bacillus, Achromobacter, Pseudomonas, Cytophaga, Sporocytophaga, Lactobacillus, Vibrio | |
A | Streptomyces | |
Lignin | F | Clavaria, Clitocybe, Collybia, Flammula Hypholoma, Lepiota, Mycena, Pholiota, Arthrobotrys, Cephalosporium, Humicola |
B | Pseudomonas, Flavobacterium |
| Download this lecture as PDF here |
MICROBIAL TRANSFORMATIONS OF NITROGEN
Biological availability of N, P and K is of considerable economic importance, since they are the major plant nutrients derived from the soil. Of the three, N stands out as the most susceptible one to microbial transformations. This element is the key building block of the protein molecule upon which all life is based on, it is an indispensable component of the protoplasm of plants, animals and micro organism.
Molecular N2 constitutes about 78% of the earth’s atmosphere but it is chemically inert and cannot be utilized by more living organism, plant animals and micro organism therefore depend on a source of combined N such as ammonia, nitrate or organic N compounds for their growth.
Nitrogen undergoes a number of transformations that involve organic, inorganic and volatile forms of nitrogen. A small part of the large reservoir of N2 in the atmosphere is converted to organic compounds by certain free living micro organism or by plant microbe association that makes the element available to plant growth. The nitrogen present in the proteins or nucleic acids of plant tissue is used by animals. In the animal body, the N is converted to other simple and complex compounds. Upon the death, plants and animals undergo microbial decay and organic N is released as ammonium, which is then utilized by vegetation or is oxidized to nitrate by microorganisms. The nitrate from of N is mostly used by the plants or may be lost by bacteria reduced to gaseous N2, which escapes to atmosphere, there by completing the cycle. The Nitrogen cycle mainly includes transformations such as
- Nitrogen mineralization : In which N containing organic complexes are decomposed and converted into inorganic compounds for use by plants
- N immobilization : In which N containing inorganic compounds are assimilated
N2 is acted on by certain micro organism sometimes in symbiosis with a higher plant, which can use it is as a N source for growth. The process of nitrogen fixation, results in the accumulation of new organic compounds in the cells of responsible micro organisms. The N2 thus fixed reenters general circulation when the newly formed cells are inturn mineralized.
By means of these reactions the subterranean microflora regulates the supply and governs the availability and chemical nature of N in soil.
NitrogenCycle

A – | Ammonium | E – | Immobilization |
B – | Mineralization | F – | Denitrification |
C – | Nitrification | G – | N2 Fixation (Non-symbiotic) |
D – | Nitrate reduction | H – | N2 fixation (Symbiotic ) |

I. Nitrogen mineralization
The conversion of organic N to the more mobile, inorganic state is known as nitrogen mineralization. As a consequence of mineralization, ammonium and nitrate are generated and organic N disappears. This takes place in two distinct microbiological steps.
1. Ammonification
It is the process of mineralization in which proteins, nucleic acids and other organic components are degraded by micro organism with the eventual liberation of ammonia. This is called ammonification. A part of the liberated ammonia is assimilated by the micro organism themselves. The first step in ammonication process is the hydrolysis of proteins, nucleic acids and other organic nitrogenous compounds into amino acids (proteolysis). The amino compounds are then deaminated to yield ammonia. Ammonification usually occurs under aerobic conditions while under anerobic conditions protein decomposition leads to conversion of ammonia into amines and related compounds (eg) clostridium. The anaerobic decomposition of protein called as putrefaction. These amines are subsequently oxidized in the presence of O2 to release ammonia.
Break down of nitrogenous substance is brought about by the activity of a multitude of microbial species.
Almost all bacteria, actinomycetes and fungi can bring about proteolysis and the amino acids produced are utilized for the growth of these organisms.
(2) Nitrification
The biological oxidation of ammonium salts (in soil) to nitrites and the subsequent oxidation of nitrites to nitrates is called as nitrification. i.e. the biological convention of N in soil from a reduced to a more oxidized state, called nitrification.
Nitrification occurs in two steps;
First ammonia is oxidized to nitrite.
2 NH3 + 1½ H2O2 → NO2- + 2H+H2O-Nitrosofication
This change is brought about by chemoautotrophic bacteria of the genera Nitrosomonas, Nitrosolobus, Nitrosococus, Nitrosospira. These bacteria obtain their energy requirement by the oxidation of NH4+ to NO-2. Among the nitrifiers Nitrosomonas are most important in soils.
Some heteotrophs involved
Streptomyces, Nocardia
Second step
Nitrite is further oxidized to nitrate
HNO2 + ½ O2 → HNO3.
Organisms: Nitrobacter, Aspergillus, Penicillium, Cephalosporium.
Factors influencing the growth of nitrifying bacteria in soil
Levels of ammonia and nitrite, aeration, moisture, temperature, pH and organic matter. In acid soils – nitrification is poor. Waterlogged soils – deficient in O2 – not congenial for nitrification.
3. Denitrification
The convention of nitrate and nitrite into molecular N2 or nitrous oxide through microbial processes is known as denitrification. Certain bacteria are capable of using nitrate as the terminal electron acceptor under anaerobic conditions. This is called nitrate respiration. As a consequence of nitrate respiration, NO3 is reduced to N2 gas or nitrous oxide. Denitirifcation leads to the loss of N from the soil. It depletes N, and therefore it is not a desirable reaction.
The escape of molecular N into the atmosphere is also known as volatalization.
Denitirfication occur mostly in waterlogged anaerobic soils with a high organic matter contents. Denitrification of bound nitrogen to gaseous N is mediated by numerous species of bacteria, which normally use O2 as hydrogen acceptor (aerobically) and, also use nitrates and nitrites (anerobically).
Anaerbic convertion of nitrate into molecular nitrogen is known as nitrate respiration.
Bacterial genera which bring about denitirfication Pseudomonas, Achromobacter, Bacillus, Micrococcus
2NO-3 +10 H → N2 + 4H2O+ 2OH- (or)
2NO-2 +6 H → N2 +2H2O +2OH- (or)
N2O + 2H → N2 + H2O
Since nitrates are used as a source of electron acceptor, there is a net loss of N from soil. This process is termed also as dissimilatory nitrate reduction. Many soil bacteria like.
Thiobacillus denitrificans
Oxidize S (chemoautotrophically) and also reduce nitrate to nitrogen
5S + 6 KNO3 + 2 H2O → 3N2 + K2SO4 + 4KHSO4 (or)
5 K2S2O3 + 8 KNO3 + H2O → 4N2 + 9 K2SO4 + H2SO4
General pathway of denitrification
Nitrate is first reduced to nitrite, which is then transformed to nitrous oxide (NO). The nitrous oxide is converted to N2 with N2O as an intermediate.
1 2 3 4
2 HNO3 → 2HNO2 → 2 NO → N2O → N2
The enzymes involved
1. Nitrate reductase 3. Nitric oxide reductase
2. Nitrite reductase 4. Nitrous oxide reductase
- Fallow soils flooded with water are more congenial for denitrification than well drained and continuously cropped soils.
- Though it is a undesirable reaction in point of view of plant nutrition, but have ecological importance. Because with out denitrification the supply of N on the earth world have got depleted and NO3 would have accumulated.
- High concentration of NO3 are toxic, denitrification is a mechanism by which some of the N is released back to the atmosphere.
5. Nitrate reduction
The reverse of nitrification process. That is the reduction of nitrate to nitrite and then ammonia. Since organisms are able to obtain cellular Nth ammonia assimilation, the process is called as assimilatory nitrate reduction.
HNO3 + 4H2 → NH3 + 3H2O
II. Nitrogen immobilization
The process of microbial assimilation of inorganic nitrogen is referred as immobilization. In contrast to mineralization, microbial immobilization leads to the biosynthesis of the complex molecules of microbial protoplasm from ammonium and nitrate. Immobilization results in a marked depression of nitrogen uptake by the plant.
The mineralization of organic N and the microbial assimilation of inorganic ions proceeds simultaneously.Both mineralization and immobilization take place regardless of the % of N in the organic N in organic matter. On the death of micro organism, the immobilized N is however released through mineralization. It is also a loss of nitrogen. NO3 when accumulated in microbial protoplasm it is referred as assimilatory NO3 reduction.
MICROBIAL TRANSFORMATION OF PHOSPHORUS AND SULPHUR
I. Phosphorus cycle (Video)
Phosphorus is only second to N2 as an inorganic nutrient required by both plants and micro organisms. Phosphate constitutes nearly 0.1% of the earth’s crust. They occur in soil in inorganic and organic forms.
The inorganic forms are derived from parent rocks or through fertilizers application and manuring with bone meal. They are soluble in water when present as phosphates of Na, K, Ca, Mg etc.
The organic phosphorus containing compounds are derived from plants and micro organisms and are composed of nucleic acids, phospholipids, lecittin, phytin and related compounds.
- Phosphorus in phytin, phospholipids and nucleic acids is found as phosphates
- Phytin is the calcium – magnesium salt of phytic acid
- Phospholipids are compounds in which phosphate is combined with a lipid, contained 10% of cell phosphorus.
- Inorganic polyphosphates are quite abundant in certain fungi
- In soil, from15-85% of the total P is organic. Soils rich in organic matter contain abundant organic P.
- Ratios of organic C to P of 100 to 300:1 N: organic P = 5 to 20: 1
In cultivated soil P present in abundant about 1100 kg/ha but most of them as not available to plants; only about 1% of the total P is in available form.
![]() PO43- in rocks and in cells
PO43- in rocks and in cells
 Acid from Thiobacillus
Acid from Thiobacillus
Microorganisms bring about a number of transformations of the element.
- Altering the solubility of inorganic compounds of P
- Mineralization of organic compounds with the release of inorganic phosphate
- Converting the inorganic, available anion into cell components, an immobilization process (analogous to that occurring with N)
- Bringing about an oxidation or reduction of inorganic P compounds
Particularly, important to P cycle are the microbial mineralization and immobilization reactions.
(1) Solubilization of inorganic phosphorus
Insoluble inorganic compounds of P are largely unavailable to plants, but many micro organisms can bring the PO4 into solution. P solubilizing are 105 to 107 / g soil.
Eg: Pseudomonas striata, Microoccus Bacillus sp., Fusarium, pergillus sp, Solubilises calcium salts, iron, aluminum, magnesium manganese phosphate.
- P is solubilized by the production of organic acids. The acids convert Ca3 (PO4)2 to di and monobasic phosphates and releases P to plants.
- Solubilization of phosphates by plant roots & micro organism is dependent on soil pH. In neutrals and alkaline soils having a content of calcium, precipitation of CaPO4 takes place. Micro organism and plant root readily dissolve such PO4 and make them available to plants.
- On contrary, acid soils are generally poor in Ca ions and phosphates and precipitated in the form of ferric or aluminum compounds which are not soluble. There, it is solubilized by the addition of PO4 solublizing micro organism.
- Phosphorus exists mainly as apatides, with the basic formula M10 (PO4)6 X2. Commonly the mineral (M) is Ca, less often Al or Fe. The anion (X) is either F- or Cl- or OH- or CO2-3. Diverse combinations of M and X results in 200 forms of P.
(2) Mineralization of organic phosphorus
Organic form of P is the larger reservoir of P in soil. By the action of bacteria, fungi and actinomycetes, bound element in remains of the vegetation and in soil organic matter is made available to succeeding generations of plants.
Among the organic phosphours compounds, lecithin, nucleic acids and phytin occupy a prominent place. Lecithin contains 9.39 % P2O5, 1.6% N and 65.36% C.
It is a process of convention of organic forms of phosphorus into inorganic available forms of P a highly significant correlation is observed between the rates of N and P convention to inorganic forms.
- Mineralization is favoured by warm temperature, with the thermophilic range being more favourable than mesophilic range.
- Neutral pH increases PO4 release, which favours microbial metabolism
- Quantity of substrate ie presence of organic P. If more P, more of mineralization
- Mineralization is mediated by the enzymes called phosphatases. These enzymes cleave phosphorus from more frequently encountered organic substrates.
- Phytases liberates PO4 from phytic acid or its Ca-Mg, Salt, Phytin. They remove PO4-s, one at a time, yield penta – tetra, di- and mono PO4 and then finally free inositol.
- Bacillus, Pseudomonas, Aspergillus, Penicillium, Rhizopus can synthesize this enzyme. Mycorrhizal (fungi) are also able to mineralize the organic forms of P and increases P uptake by the plants.
(3) Immobilization
Process of assimilation of P into microbial nucleic acids, phospholipids or other protoplasmic substances is called immobilization. It leads to the accumulation of non utilizable forms of the element.
P accounts for 0.5-1.0% of fungus mycelium and 1.0 to 3.0% of the dry weight of the bacteria and actinomycetes.
(4) Oxidation reduction reactions
Biological oxidation of reduced phosphorus compounds into oxidized state.
Phosphite (HPO3=) is oxidized to phosphate. A number of hetertrophic (bacteria), (fungi) & (actinomycetes) utilize phosphite as sole P source. Hypophosphites (HPO2=) can also be oxidized to phosphate by heterotrophs.
HPO3= → HPO4=
HPO2= → HPO4=
Reductive process, reductive pathway has also been functioned. PO4 is reduced to phosphite and hypophosphite.
H3PO4 → H3PO3 → H3PO2
Clostridium butyricum, E. coli form phosphite and hypophosphite from orthophosphate. It is biochemically analogue to the process of denitirification. Only little information is available about this process.
P exist in an organic form in the protoplasm on the death of living organism, this (P) is changed to inorganic phosphoric acid. This is soon converted into insoluble salts of Ca, Fe, Mg and Al. Phosphorus thus alternates between organic and inorganic, and soluble and insoluble forms. In soluble P is solubilized by various acids produced by micro organism.
Microbial activities involved in the cycling of C, N and P are absolutely essential for maintenance of soil fertility.
II. Sulphur cycle
Sulphur like N, is an essential element for all living systems because of it’s inert nature, is not utilized by plants. To be used first S has to to be oxidized or reduced. In soil, it occurs both organic (S containing amino acids, vitamins ) as well as inorganic forms (Sulphur, sulphates etc., ) .
Four distinct transformations are recognized
- Decomposition/Mineralization of larger organic S compounds to smaller units and their conversion into inorganic compounds
- Microbial associated immobilization
- Oxidation of inorganic ions and compounds such as sulphides,thiosulphates, Sulphu
- Reduction of Sulphates abd other sulphides

 Mineralization
Mineralization
Conversion of organic bound S into inorganic state, mediated through M.O. The released S in either absorbed by plants or escaped into atmosphere in the form of oxides
Oxidation
- Occurs both in aerobic and anaerobic condition
- Bacteria
- Nonfilamentous forms- Thiobacillus
- Filamentous forms – Beggiatoa,Thiothrix and Thioloca
- Fungi and actinomycets
- Aspergillus, Penecillium and Microsporium
Importance of Thiobacillus
- Produces Sulphuric acid ,lower down the soil pH – Hence used in controlling plant disease
- Apple and Potato scab –Streptomyces scabis ,Sweet potato rot – S. ipomea
- S+ Thiobacillus application is used for the control
- Remediation of alkali soil
- Increases the solubilization of other nutrients (P,K,Ca,Mn,Al and Mg )
- Preparation of biosuper- Rock phosphate + T.thiooxidans and S— Australia
- Lipman’s process- Compost preparation
- Soil + manure + elemental S + rock phosphate
Sulphate reduction
Reduces inorganic sulphate into Hydrogen sulphide –reduces the availability of S for plant nutrition
- Desulphovibrio desulphricans -anaerobe
| Download this lecture as PDF here |
Fixation of elemental nitrogen in the atmosphere by the micro organism through a reductive process into ammonia is called as BNF. A variety of prokaryotic organism have the ability to reduce the atmosphere N2 BNF accounts for about 70% of the total N fixed in the biosphere. The ability to reduce atmosphere N is restricted only to bacteria, which are belonging to the diverse groups. The root nodule associations were the first to be recognized for their ability to fix atmosphere N2. Rhizobia are the first group of organism realized for its potential of nitrogen fixation.
Nitrogen fixing bacteria
Nitrogen fixing bacteria are classified according to their mode of fixation.
- Free living N fixers – capable of fixing mol. N2 to cellular N independently of other living organism.
- Associative N fixers
- Endophytic N fixers
- Symbiotic N fixers
Rhizobium is predominant symbiotic N2 fixing bacterium. Boussingault showed that leguminous plant can fix atmosphere N2. Then hellriegel and wilfarth – proved that N2 is fixed by certain bacteria living in root nodules of leguminous plants. Latter isolated in pure culture by Beijerinck. Winogradsky isolated clostridium pasteurianum. Which is an anaerobic N2 fixer. Beijerinck isolated Azotobacter as free living aerobic N2 fixing organisms.
Cross inoculation groups of rhizobium (CIG)
It (CIG) refers the groups of leguminous plants that will develop effective nodules when inoculated with the rhizobia obtained from the nodules from any member of that legume group.
I. Rhizobium
| 1. | Rhizobium leguminosarum | CIG | Host it can nodulate |
| bv. viceae | Pea | Peas,lenfils, vicia |
| bv. phaseoli | Bean | Phaseolus spp |
| bv. trifoli | Clover | Trifolium spp |
2. | R. meliloti | Alfalfa | Alfalfa, clover, fenugreek |
3. | R. loti | Lotus | Trifoli, lupine, |
4. | R. fredii | Soybean | Soybean |
5. | R. spp | Cowpea group | Vigna, Arachis, Cajanus, Dolichus, Sesbania, Acacia, Prosopis, green gram and blackgram |
6. | R. sp | Chickpea group | Chickpea |

II. Bradyrhizobium
- B. japonicum Soybean
- B. spp Cowpea group
III. Azorhizobium – Stem nodulating – one. Nodulates Sesbania rostrata.
IV. Photorhizobia – Nodulants aeschynomene sp.
V. Sinorhizobium – fast growing soybean nodulator
I. Biological nitrogen fixation
Free living nitrogen fixers
- Azotobacter – Aerobic
- Beijerinckia
- Clostridium – Anaerobic
- Cyanobacteria (Blue green algae) etc.,
II. Associative symbiotic nitrogen fixer
Azospirillum
Herbaspirillum
III. Endophytic nitrogen fixer
Gluconacetobacter diazotrophicus
IV. Symbiotic nitrogen fixers
- Rhizobium (Rhizobium – legume association)
- Bradyrhizobium (Bradyrhizobium – soybean association)
- Azorhizobium (Azorhizobium- Sesbania rostrata association)
- Anabaena azollae (Azolla – Anabaena association)
- Frankia (Frankia – Casuarina association)
Species of Azospirillum
- lipoferum
- brasilense
- amazonense
- halopareferans
- irkense
- A.largomobilis
Species of Azotobacter
- chroococcum
- vinelandii
- beijerinkii
- paspali
- agilis
- insignis
- macrocytogens
Important genera of blue green algae
Anabaena, Nostoc, Cylindrospermum, Rivularia, Oscillatoria, Plectonema, Aphanothece, Lyngbya, Scytonema, Calotrhix etc.,
Species of Azolla
- pinnata
- filiculoides
- microphylla
- caroliniana
- mexicana
- nilotica
Nitrogen fixation
Process of N2 fixation
The process of N2 fixation is mediated by the enzyme, called nitrogenase (which mediates the reduction of N2 to ammonia) first, this enzyme was extracted from the anaerobic di nitrogen fixer Clostridum pasteurianum. Latter, this has been isolated form most other N2 fixing bacteria.
The mechanism of N2 fixation appears to be quite similar in most N2 fixing prokaryotes. The enzyme has been fairly well characterized and the enzymes from these different systems share common properties allowing a unified single doscription of nitrogenase.
Nitrogenase
Nitrogeanse is a functional enzyme which reduces N2 to ammonia and depends on energy source from ATP. The nitrogenase has two components one containing Mo-Fe, designated as Mo – Fe protein and the other Fe protein . Two components are necessary for the nitrogenase activity.
Mo-Fe protein
Consists of 4 subunits and having the molecular not of 22,0000 or 270,000 daltons and it is the big component.
Fe-protein
Smaller component, contains 2 subunits, molecular weight 60,000 daltons.
Ammonia is the end product of N2 fixation. The over all reaction is as follows.
ATP![]() N2 + 3H2 2 NH3
N2 + 3H2 2 NH3
General pathway of N2 fixation
This process requires a source of ATP and reductants, which are provided by photosynthesis. 16 molecules of ATP are required to fix a molecule of N2.
|
|
|
|
| ADP |
|
|
|
|
| N=N |
|
| Electron carriers |
|
|
|
|
|
|
|
| NH3 |
Nitrogenase can also reduce C2 H2 → C2H4
Hydrolysis of ATP into ADP with electron transfer from a reduced electron donor (Ferridoxin, Flavodoxin) is coupled to reduce N2 to 2NH3. The ammonia is the first stable product of fixation and it is assimilated by GS-GOGAT pathway.
| NADH | |||||||||
| NADPH | e- |
| e- |
| e- |
| e- | ||
Ferridoxin | Flarodoxin | ||||||||
| APP | |||||||||
| EMP | e- Carrier proteins | NII | NI | ||||||
| EDP | |||||||||
| Glycolysis | |||||||||
Nitrogenase is O2 labile various protection mechanism are operating in different N2 fixing systems.
![]()

Mechanism
Reduction takes place on the surface of the enzyme
- Six electrons are required to reduce one mole of N to two moles of ammonia.
N2 + 8H+ +8e- +16 ATP ——-2 NH3+16 ADP+ 2H+ +16pi

It is postulated that, atoms of N2 are separated thr’h charge in the valency of metal ion (mo) bound to the enzyme involved in reduction of N2. For every electron transfer, 4 ATP mole are required.
Hydrogenase –Uptake hydrogenase (HUP+) converts the release d hydrogen during N2 fixation, and cycled back the Hydrogen for energy generation.by this they contribute 9-10 %ATP requirement for N2 fixation process.
Formation of a Root Nodule
Factors affecting N2 fixation
- Presence of nitrate or ammonium : More N2, No, N2 fixation
- Presence of certain inorganic substances
Ca, Co, Mo – influence N2 fixation along with P
- Availability of energy source – addn. of C source increase N2 fixation
- pH : Neutral – favours Azotobacter – Acidic- Beijerinkia
- Soil moisture : Adequate is good for fixation
- Temperature: Mesophilic – 30°C.
The energy requirement for BNF is very high and it is a major factor determines the amount of N2 fixed. In, Azotobacter the rate depends on amount of available carbon. In symbiotic N2 fixers since photosynthesis is the ultimate source of energy the rate of N2 fixation is influenced by the factors that effect photosynthesis and rate of translocating photosynthates to the N2 fixing system.
Nitrogenase protection mechansims
- Leghaemoglobin scavenges O2 to protect nitrogenase in legume rhizobium symbiosis
- Confirmatory protection in Azotobacter as well as the higher respiratory rate.
- Thick walls of Heterocyst protect O2 in BGA, since Nitrogenase are present in the heterocyst.
- Microaerophilic nature in Azospirillum
Losses of N by non biological ways
Leaching
20 to 50%of fertilizer N. The most striking loss of N in rice soils where more than half of the fertilizer N applied get lost through leaching.
Volatalization
Another factor is the volatalizaiotn of ammonia in soil 5-20%.
Fixation of ammonium in soils is the minor contributory factor to overall loss of N2 available for plant growth.
Such losses of N by physical causes and by nitrification and denitirfication process can be controlled by the application of certain chemicals. Some chemicals have been designed to control the rate of release of nutrient from nitrogenous fertilizers, while others retard nitrification in soil by controlling the activity of nitrifying bacteria.
a. Controlled release fertilizers
| Urea from… isobutyeldene diurea Crotonilidene diurea S coated urea | Fertilizers, sparingly soluble in water can regulate the release of N from fertilizers |
b. Nitrification inhibitors
These are substituted with pyridines, pyrimidines, anilines and isothiocyanates,
Examples
1. 2 chloro 6 (tricholormethyl) – pyridine – (N serve )
2. 2 amino 4 chloro 6 methyl pyridine –( AM.)
N serve inhibits the growth of Nitrosomonas europea and N. agilis.
The seeds of neem conain lipid associates act as nitrification inhibitors and there by increases the efficiency of urea fertilizers.
Ammonia assimilation
N2 fixation results in NH4 formation which reacts with organic acids and form amino acids which is mediated by ammonia assimilating enzyme.
GS – Glutamine synthetase
GOGAT – Glutamate synthese
GDH – Glutamate dehydrogenase
Genetics
Nif genes are responsible for N2 fixation.
Nif genes are 22, which are located in 7 or 8 clusters.
GENETICS OF NODULATION AND N FIXATION
Root nodule bacteria and symbiosis with legumes
One of the most interesting and important plant bacterial interactions is that between leguminous plants and certain gram negative nitrogen fixing bacteria. Rhizobium, Bradyrhizobium, Sinorhizobium, Mesorhizobium and Azorhizobium are gram negative motile rods. Infection of the roots of a leguminous plant with the appropriate species of one of these genera leads to the formation of root nodules that are able to convert gaseous nitrogen to combined nitrogen, a process called nitrogen fixation. Nitrogen fixation by legume Rhizobium, symbiosis is of considerable agricultural importance, as it leads to very significant increases in combined nitrogen in the soil. Because nitrogen deficiencies often occur in unfertilized bare soil, modulated legumes are at a selective advantage under such conditions and can grow well in areas where other plants cannot.
- CIG refers, the groups of leguminous plants that will develop effective nodules when inoculated with the rhizobia obtained from the nodules from any member of that legume group.
Stages in root nodule formation
The stages in the infection and development of root nodules are not fairly well understood. They include
- Recognition of the correct parameter on the part of both plant and bacterium and attachment of the bacterium to the root hairs.
- Excretion of nod factors by the bacterium.
- Invasion of the root hairs by the bacterial formation of an infection thread.
- Travel to main root via the infection thread.
- Formation of deformed bacterial cells, bacteroids, within the plant cells and development of the nitrogen fixing state.
- Continued plant and bacterial division and formation of the mature root nodule.

Nodulation events
- Normal root hair
- Exudation of organic substances
- Accumulation of rhizobia in the rhizosphere
- Orientation and binding of rhizobia
- IAA production
- Root hair curling and deformation
- Formation of infection thread by rhizobia
- Formation of sheperd’s crook cells and entry of infection thread
- Thread containing bacteroids extending into root hair cells
- Entry of infection thread into cortex and branching
- Nodule development
Attachment and infection
The roots of leguminous plants secrets a variety of organic compounds that stimulate the growth of a rhizosphere micro flora. This stimulation is not restricted to the rhizobia but occurs with a variety of rhizosphere bacteria. If there are rhizobia in the soil, they grow in the rhizosphere and build up to high population densities. Attachment of bacterium to plant in the legume Rhizobium symbiosis is the first step in the formation of nodules. A specific adhesion protein called rhicadhesin is present on the surface of all species of Rhzobium and Bradyrhizobium. Rhicadhesin is a calcium-binding protein and may function by binding calcium complexes on the root hair surface. Other substances, such as carbohydrate-containing protein called lectins, also play in plant bacterium attachment.
Initial penetration of Rhizobium cells into the root hair is via the root hair tip. following binding, the root hair curls as a result of the action of substances excreted by the bacterium called nod factor and the bacteria enter the root hair and induce formation by the plant of a cellulosic tube, called infection thread, which spreads done the root hair. Root cells adjacent to the root hairs subsequently become infected by rhizobia and nod factors stimulate plant cell division, eventually leading to formation of the nodule.
Bacterioids
Bacteriods are specifically referred to a swallon deformed Rhizobium cellfound in the root nodule,capable of nitrogen fixation
The Rhizobium bacteria multiply rapidly with in the plant cells and are transformed into swollen, misshapen and branched forms called bacteroids. When the plant dies, the nodules can be deteriorates, releasing bacteria into the soil. The bacterioid forms are incapable of division, but there are always a small number of dormant rod shaped cells present in the nodule. These now proliferate; using some of the products of the deteriorating nodule as nutrients, and the bacteria can initiate the infection in other roots or maintain a free living existence in the soil.
Lectins
Plant proteins which specifically bind to carbohydrate receptors (polysaccharides) in the rhizobial cell
Genetics of nodule formation
Genes directing specific steps in nodulation of a legume by a strain of Rhizobium are called nod genes. Many nod genes from different Rhizobium species are highly conserved and are borne on large plasmids called sym plasmids. In addition to nod genes which direct specific nodulation events, sym plasmids contain specificity gene, which restrict a strain Rhizobium to a particular host plant. Indeed cross inoculation group specificity can be transferred across species of rhizobia by simply transferring the respective sym plasmid.
In the sym plasmid of Rhizobium leguminosarum bio var viciae, nod genes are located between two clusters of genes for nitrogen fixation the nif genes. Ten nod genes have been identified in this species. The nod ABC genes are involved in the production of oligosaccharides called nod factors, which induce root hair curling and trigger plant cell division, eventually leading to formation of the nodule. In Rhizobium leguminosarum bio var viciae, the gene nodD encodes a regulatory protein; this controls transcription of other nod genes.
Nod D genes
- Genes directing specific steps in nodulation of a legume by a strain of rhizobium are called Nod genes
- Nod genes are born on large plasmids ,called sym plasmids
- Nod genes are located between two clusters of genes for N2 fixation called Nif genes
- Nod gene consists of 8 genes
- nod A,B,C,D,E,F,L,M
- nod D controls the function of all nod genes
Nif Genes
- Genes responsible for N fixation are called Nif genes
- 22 genes are involved, arranged in 7 /8 clusters
- Nif Q,B,A,L,F,M,Z,W,V,S,U,X,N,E,Y,T,K,D,H,J
- KDH – control Nitrogenase enzyme complex
Factors affecting nodulation
- Temperature and light
- Combined Nitrogen
- Hydrogen iron concentration
- Mineral nutrition-Co,Mo,P,Ca
- Genetic factors
- Ecological factors
- Salinity and alkalinity
| Download this lecture as PDF here |
Aerial plant surfaces represent the largest biological interface on Earth and provide essential services as sites of carbon dioxide fixation, molecular oxygen release, and primary biomass production. Rather than existing as axenic organisms, plants are colonized by microorganisms that affect both their health and growth.
For terrestrial plants, the phyllosphere represents the interface between the above-ground parts of plants and the air. Conservative estimates indicate that the roughly 1 billion square kilometers of worldwide leaf surfaces host more than 1026 bacteria, which are the most abundant colonizers of this habitat . The overall microbiota in this ecosystem is thus sufficiently large to have an impact on the global carbon and nitrogen cycles. Additionally, the phyllosphere inhabitants influence their hosts at the level of the individual plants. To a large extent, interest in phyllosphere microbiology has been driven by investigations on plant pathogens. Their spread, colonization, survival, and pathogenicity mechanisms have been the subject of numerous studies. Much less understood are nonpathogenic microorganisms that inhabit the phyllosphere. The composition of the phyllosphere microbiota has been analyzed in only a few studies by cultivation-independent methods; however, such methods are essential in light of the yet uncultivated majority of bacteria existing in nature, or more specifically on plant leaves. Not only their identity, but in particular the physiological properties of phyllosphere bacteria, their adaptations to the habitat, and their potential role (e.g., with respect to modulating population sizes of pathogens) remain largely unknown. Current knowledge on the traits important in the phyllosphere is derived from relatively few studies on gene expression and stems mostly from model bacteria cultivated on host plants under controlled conditions. However, under natural conditions, plants and their residing microorganisms are exposed to a host of diverse, highly variable environmental factors, including UV light, temperature, and water availability; moreover, individual microbes are subjected to competition with other microorganisms over resources, such as nutrients and space.
Toward a deeper understanding of phyllosphere microbiology, and in particular to learn more about the commensal majority of plant leaf colonizing bacteria, which may be of relevance for plant health and development, integrated approaches are needed. Here,
Bacterial communities in the phyllosphere are thought to be limited by carbon availability, and it may be expected that access to carbon compounds on leaves is a major determinant of epiphytic colonization. There is evidence that small amounts of nutrients, such as simple sugars including glucose, fructose, and sucrose, leach from the interior of the plant.
The above-ground parts of plants are normally colonized by a variety of bacteria, yeasts, and fungi. While a few microbial species can be isolated from within plant tissues, many more are recovered from the surfaces of healthy plants. The aerial habitat colonized by these microbes is termed the phyllosphere, and the inhabitants are called epiphytes. While there has been some investigation of the colonists of buds and flowers, most work on phyllosphere microbiology has focused on leaves, a more dominant aerial plant structure. Bacteria are by far the most numerous colonists of leaves, often being found in numbers averaging 106 to 107 cells/cm2 (up to 108 cells/g) of leaf. Because of their numerical dominance on leaves, and because more information is available on the process of bacterial colonization of leaves, we focus on this group of microbes in this review.
Compared to most other bacterial habitats, there has been relatively little examination of phyllosphere microbiology. This is somewhat surprising given the abundance of plants in the world and the roles of various phyllosphere bacteria in the important processes discussed below. Leaves constitute a very large microbial habitat. It is estimated that the terrestrial leaf surface area that might be colonized by microbes is about 6.4 x 108 km2. Given the large number of bacteria on leaves in temperate regions of the world and that populations in tropical regions are probably even larger, the planetary phyllosphere bacterial population may be as large as 1026 cells. Clearly, in aggregate, these bacteria are sufficiently numerous to contribute in many processes of importance to global processes, as well as to the behavior of the individual plants on which they live.
The microbial communities of leaves are diverse and include many different genera of bacteria, filamentous fungi, yeasts, algae, and, less frequently, protozoa and nematodes. Filamentous fungi are considered transient inhabitants of leaf surfaces, being present predominantly as spores, whereas rapidly sporulating species and yeasts colonize this habitat more actively. Bacteria are by far the most abundant inhabitants of the phyllosphere. Epiphytic bacterial populations differ sharply in size among and within plants of the same species, as well as in close proximity, and over short time scales as well as over the growing season. These considerable variations in population sizes are caused in great part by the large fluctuations in the physical and nutritional conditions characteristic of the phyllosphere. Additionally, plant species appear to influence the microbial carrying capacity of the leaf, since the total number of culturable bacteria recovered from broad-leaf plants such as cucumber and beans was significantly greater than that recovered from grasses or waxy broad-leaf plants.
Reflective of marked differences in the physicochemical environments of above-ground versus subterranean plant surfaces, the leaf bacterial flora differs substantially from that of roots. For example, pigmented bacteria, which are rarely found in the rhizosphere, dominate leaf surfaces, presumably because solar radiation influences the ecology of the phyllosphere. The differential composition of leaf and root bacterial communities is further evidenced by the failure of common root colonizers such as Rhizobium and Azospirillum to become established on leaves.
Studies of the composition of bacterial communities on leaves have been numerous but rather limited in scope. It is generally believed that populations of culturable aerobic bacteria on leaves are dominated by a few genera. A few exhaustive studies of the variations in the microbial community of leaves over multiple time and space scales have provided important detailed knowledge about the identity and the ecology of bacterial leaf inhabitants. Ercolani made an extensive inventory of culturable aerobic bacteria isolated from the surface of olive leaves over six growing seasons and reported distinct bacterial community structures on leaves of the same age at a given time of the growing season. Thompson et al. analyzed 1,236 bacterial strains from immature, mature, and senescent leaves of field-grown sugar beets over a complete growing season. They identified 78 species and 37 named and 12 unnamed genera of bacteria. Most importantly, like Ercolani, they found distinct patterns of microbial colonization at different times of the year, with bacterial community diversity being lowest during the warmest and driest months of the season and highest during the cooler and rainy months. Coincidentally, in both of the above-described studies, communities on young leaves were composed of a greater number of taxa than those of old leaves. Thus, specific natural environments of the phyllosphere apparently select for the presence of specific genotypes within the leaf bacterial community. This is further supported by the finding that the acquisition by Pseudomonas fluorescens of plasmids that are indigenous to the leaf microflora coincided with a specific maturation stage of the plant over two consecutive years. This indicated that traits carried on these plasmids conferred variable selective fitness to specific plasmid-bacterial host combinations during the growing season, possibly in response to changing conditions in the phyllosphere habitat.
The study of bacterial colonizers of leaves has been restricted mostly to aerobic culturable bacteria and also driven by the importance of investigating the ecology of plant-pathogenic bacteria because of their deleterious effect on plant productivity. Thus, the microbial ecology of the phyllosphere has been viewed mainly through the biology of gram-negative bacteria such as Pseudomonas syringae and Erwinia (Pantoea) spp., two of the most ubiquitous bacterial participants of phyllosphere communities. There is reason to believe, however, that the extreme fluctuations in the physicochemical environment of the phyllosphere over short time scales may select for bacterial species that have unusual and versatile traits that make them fit to colonize plant surfaces but have remained unculturable. The leaf surface has long been considered a hostile environment for bacterial colonists. The leaf surface is exposed to rapidly fluctuating temperature and relative humidity, as well as repeated alternation between presence and absence of free moisture due to rain and dew. The leaf also provides limited nutrient resources to bacterial colonists. While other habitats probably offer more extreme conditions of desiccation or temperature, etc., they may not be subject to such rapid and extreme fluctuations in these several physical conditions. Several factors may influence the microhabitat experienced by bacteria on leaves. First, the leaf itself is surrounded by a very thin laminar layer in which moisture emitted through stomata may be sequestered, thereby alleviating the water stress to which epiphytes are exposed. Second, some cells in a leaf bacterial population, particularly in plant-pathogenic populations, may not reside in exposed sites on the leaf surface but instead may at least locally invade the interior of the leaf, avoiding the stresses on the exterior of the leaf by residing in substomatal chambers or other interior locations. Thus, while some phytopathogens may have the option of avoiding stresses, most other epiphytes apparently must tolerate them in some way.
The phyllosphere has many features that make it an excellent habitat in which to study microbial ecology. Leaves are clean, and microbes can be observed directly on leaves, enabling the use of powerful new microscopic techniques to measure microbial identity, activity, and gene expression. Plants can be readily grown without epiphytic microbial communities, allowing us to readily manipulate their inhabitants, while communities can be made as simple or complex as needed by simple inoculation. In addition, important microbial processes, such as immigration, and ecological models, such as island biogeography, can be readily explored in epiphytic bacterial systems. Thus, phyllosphere microbiology has much to offer to the field of microbial ecology and promises to contribute to more effective and less environmentally damaging means of plant protection.
| Download this lecture as PDF here |
Composting is the active process of converting organic material to more stabilized forms of C through the action of microorganisms. Specifically, composting is the biological decomposition of wastes consisting of organic substances of plant or animal origin under controlled conditions to a state sufficiently stable for storage and utilization (Diaz et al., 1993). Compost as a product can be used in gardens, in nurseries, and on agricultural land. With respect to management of organisms, composting is perhaps the prime example since we manage the microbial process and the microbial product and manage the use of compost in microbially based systems (Cooperband, 2002). As the compost definition implies, practically any plant or animal material can be composted. Compost plays a major role in the agriculture of developing countries using organic agriculture and biodynamic farming, being relied upon to provide organic matter and nutrients and increase soil tilth. It also plays a role in processing the human waste stream. The United States alone produces nearly 10 MMT of sewage sludge and 185 MMT of garbage annually, on a dry weight basis. Less than 15% of municipal solid waste is recycled; however, more than 30% of the sewage sludge is beneficially used as composted products (Rynk, 1992; http://compost.css.cornell.edu/OnFarmHandbook).
Traditionally yard waste is thought of as “the” compost material; however, manure, meat and dairy waste, wood, sawdust, and crop residue can be composted. In addition, animal carcass composting is receiving significant attention due to the environmental benefits versus burial, which can contribute to groundwater contamination.
One important aspect of the material that affects the compost process and product is the C:N ratio of the starting material, ideally it should be 25 to 30:1. Typical C:N ratios of different materials are shown in Table below:
C:N Ratios of Various Compost Materials
| Materials | C:N |
Activated sludge | 6 |
Grass clippings | 12–15 |
Manure | 20–50 |
Poultry manure | 15 |
Soil humus | 10 |
Sawdust | 200–500 |
Vegetable waste | 12 |
Wheat straw | 80 |
Wood | 400 |

The organisms and processes occurring during composting of straw. The length of time varies with outside temperature and extent of mixing but usually involves 200 days (from W. R. Horwath, personal communication).
Materials can be mixed to adjust the C:N ratio for a consistent product. There are numerous methods used for preparing materials and the environment for the composting process, including using waste materials alone, mixing organic materials of different quality, adding external nutrients and/or inocula, and controlling the physical environment to promote aerobic or anaerobic decomposition. In compost terminology, process strategy refers to the management of the biological and chemical activity of the composting process. The biological processing uses terminology referring to the stage of the composting, such as active stage (mesophilic), high-rate stage (thermophilic), controlled (cooling), and curing stage (maturing) (Fig. 17.2). Configuration refers to the physical management of the process such as using piles, stacks, or windrows. Composting configurations range from windrow or open systems to enclosed systems, with windrow further classified as either static or turned. An example of a static system would be a stationary undisturbed mound of organic material with air being forced up through the mound or pulled down through the mound. In contrast, a turned system uses mixing as the aeration method, which also enhances the uniformity of decomposition and reduction in material particle size. The turned system is considered the traditional composting method for organic material (see Diaz et al., 1993, for more detail). With any composting system managing the composting process will consistently produce compost with the desired characteristics.
The most prevalent composting technique is aerobic decomposition, carried out by a diverse microbial population that changes composition as conditions change. This method is preferred since it proceeds more rapidly and provides a greater reduction in pathogens and weed seeds because higher temperatures are achieved. Physiochemical factors affecting aerobic composting are temperature, moisture, aeration, pH, additives, particle size, and the C:N ratio of the composted substrate. Mostly indigenous organism populations are used for the composting process; microbial inoculants are utilized only under certain conditions. Figure 17.2 depicts the process of composting straw for 200 days under optimum conditions of temperature and moisture. In the mesophilic stage metabolism of the labile-C-rich substrates increases rapidly, generating heat. At this point there is a mixture of bacteria, actinomycetes, and fungi contributing to the decomposition process. In the early and transition stage to thermophilic conditions the windrow is turned, causing a decline in temperature and oxygenation of the inner material, resulting in rapid decomposition and temperature increase. As the temperature reaches 40°C the system turns from a mesophilic to a thermophilic stage, favoring mainly thermophilic bacteria and actinomycetes, with Bacillus being the dominant genus. Common Bacillus species found at this stage, and accounting for 10% of the decomposition, are brevis, circulans, coagulans, licheniformis, and subtilis. Decomposition will continue in the thermophilic zone until substrates begin to decline, then a gradual decrease in temperature will occur.
As the temperature declines the mesophilic organisms reappear, especially fungi that have preference for the remaining lignin and cellulose substrates. Fungi, responsible for 30 to 40% of the decomposition the compost material, include Absidia, Mucor, and Allescheria spp., haetomium, thermophilum, dactylomyces. The actinomycetes, such as the Norcardia spp., Streptomyces thermofuscus, and S. thermoviolaceus are important in this phase when humic materials are formed from decomposition and condensation reactions. The actinomycetes are estimated to account for 15 to 30% of the decomposition of composted material.
The compost produced from this process is lower in C than the initial material, has a lower C:N and a higher pH, and can contain considerable NO3. The end product of composting depends on the original substrate, any added nutrients, degree of maturity, and composting method; typical properties of composted plant material are listed in Table 17.4. Adding compost to soil increases the SOM, which increases soil structure and water-holding capacity and infiltration. In addition, compost contains significant amounts of plant nutrients such as N, P, K, and S and micronutrients, which are slowly released into the soil. As an ancillary benefit compost contains fairly resistant C compounds and may be dominated by fungi. Using compost on a garden or agricultural soil would favor an increase in the population of fungi and thus an increase in the fungi:bacteria ratio. Fungi are very abundant in soils and can constitute as much biomass as roots and as a group they are also the major organic matter decomposers in soil. Increasing the soil fungal population can increase C compounds that are agents in binding soil particles into aggregates, which increase soil tilth. Recent studies (Bailey et al., 2002) haveshown there is increased soil C storage in soils with greater fungal:bacterial ratios. Thus, as a consequence of using compost on our soil we have managed the soil microorganism population to our benefit.
General Compost Properties
% N | >2 |
C : N | <20 |
% Ash | 10–20 |
% Moisture | 10–20 |
% P20 | 15–1.5 |
Colour | Brown black |
Odour | Earthy |
% Water-holding capacity | 150–200 |
CEC (meq 100 g–1) | 75–100 |
% Reducing sugars | <35 |
CROP ROTATIONS AND GREEN MANURES
Crop rotations have been practiced over the long history of agriculture. Studies dating from the 1840s on have shown that N supplied to grain crops was the major reason for using crop rotations containing legumes (Triplett and Mannering, 1978). With the advent of inexpensive nitrogen fertilizers, crop rotations containing legumes declined. Only recently has the value of crop rotations specifically including legumes been recognized as critical in maintaining SOM and soil productivity. Researchers in Canada studied the nutrient dynamics in a Canadian Luvisol after 50 years of cropping to a 2-year rotation (wheat–fallow) or a 5-year rotation (wheat–oats–barley–forage–forage) (McGill et al., 1986). Their results showed that the soil cropped to the 5-year rotation contained greater amounts of organic C and N. In addition they found that microbial turnover (i.e., carbon mineralization) was twice as fast in the 2-year rotation. The 5-year rotation doubled the input of carbon into the soil over the 2-year system and had a greater percentage of organic C and N in biological form. These results suggest that longer cropping system rotations that include forage or legumes will conserve SOM, maintain a greater biological nutrient pool, and put more nutrients into the soil than intensive rotations.
In a 10-year study, a low-input diverse crop system with manure and a low-input cash grain system with legumes showed significant increases in SOM compared to a conventional corn/soybean rotation (Wander et al., 1994). In addition, in both low-input (multiple crop rotations) systems the microbial biomass was greater and its activity higher than the conventional rotation of corn/soybeans with chemical inputs. The low-input systems also mineralized significantly more N and the microbial biomass contained 33 kg N ha_1 more N than the conventional system.
n agricultural systems, plant pathogens are an important part of the soil microbial community. As growers reduce tillage and incorporate a greater variety of crops in rotation they face an increasing number of plant diseases that can cause significant stand and yield reductions. These potential losses, however, may be offset in systems incorporating green manures by promoting disease-suppressing properties that reduce plant pathogens, either (1) by increasing the levels of SOM that create conditions supporting a greater microbial biomass, competition for resources, antibiosis, or antagonism or (2) through direct inhibition by production of antibacterial/fungal compounds as in the case of Brassica cover crops that produce isothiocyanates. Cover crops are known to control disease-causing organisms through competition for resources and space, control of soil micronutrient status, and alteration of root growth.
| Download this lecture as PDF here |
- Microbes are nature’s decomposers. The variety of metabolic abilities in microbes is enormous, and includes microbes that can degrade or mitigate all sorts of human products and activities, from oil spills to pesticide runoff to toxic waste.
- Environmental Microbiology seeks to find ways to maximize the efficiency of microbes in helping to remove various kinds of wastes (e.g. sewage treatment), or to minimize the opportunities of microbes to produce problems (e.g. water treatment).
- Environmental Microbiology is a growing field, often brings together issues of concern to engineers, geologists and hydrologists, microbiologists, and public health officials.
Sewage Treatment
- Until 1900’s, human wastes were simply dumped as raw sewage into the nearest outhouse, stream, or river. As connection of sewage to diseases such as cholera became clear, public policy changed to required water treatment. This had major impact on reduction of many diseases.
- Sewage = mix of domestic + industrial waste plus drainage water from rainfall. Contains many microbes, mostly harmless but some pathogens from humans or animals. Can include bacteria such as Vibrio cholera, Shigella dysenteriae, enteropathogenic strains of E. coli and B. cereus, viruses such as Hepatitis A, many more.
- Sewage treatment: goal is to get rid of pathogens, also reduce organic content of effluent to a low level.
- Primary wastewater treatment: use screens to remove large objects (plastic bags, wads of paper, etc.), then move water to large tank to allow settling of heavier particulate matter as sludge.
- Secondary wastewater treatment: modern facilities use “activated sludge process”. After moving water from primary settling tank, bubble air through a secondary tank. Aerobic microbes grow and break down organic matter in the tank. Then move water to another tank called the secondary clarifier, where solids settle and are added to sludge. Clarified liquid is treated with chlorine to kill remaining microbes, then discharged as clear liquid into nearest river. Anaerobic bacteria break down organic matter, produce lots of fermentation products. Methanogens grow on these wastes and produce methane gas as waste. This can be trapped and used as fuel (useful in developing countries).
- Tertiary wastewater treatment: Secondary wastewater treatment does not remove inorganic ions, such as NH3, PO4-3, SO4-2. Wastewater can enrich local waters to create eutrophic conditions, including algal blooms and sufficient loss of oxygen that fish die. To prevent this, a few water treatment systems use additional steps to remove ammonia and phosphate, using additional processing tanks in which specific bacteria are used to remove ammonia and phosphate.
Bioremediation
- Expanded use of chemicals in industry has produced major new problems of environmental pollution. U.S. alone has over 50,000 hazardous waste sites. Entire communities have been evacuated because of accumulated toxic wastes. Groundwaters are often polluted as well, including toxic chemicals such as commercial solvents used to degrease machinery or in “dry cleaning”. Fertilizers and pesticides are often found in water downstream from agriculture.
- Bioremediation = use of living organisms to promote destruction of environmental pollutants. See example of anaerobic toluene degrader .
- Typically, native microbes are used (rather than introduced or genetically mofified organisms). Rather than waiting for “nature to take its course”, try to speed up the process. How?
- “Pump and treat”. One way to speed bioremediation. Pump groundwater to surface, add nutrients (e.g., O2, methane), reinject into contaminated zone. In some cases, can inject
- Bioreactor. Another technique. Put contaminated soil or groundwater into an industrial-sized fermentor, add appropriate microbes to degrade materials, keep adding more substrate over time. This works well for very toxic chemicals such as chlorinated compounds (PCBs).
- Microbiology of water
Drinking water is obtained either from surface sources such as rivers, lakes or from underground water. Such natural waters are likely to be polluted with domestic and industrial wastes. Although water purification systems envisage protection from pollution, sometimes, the water supply can become a potential carrier of pathogenic organisms and endanger public health. A number of diseases such as cholera, typhoid, viral hepatitis etc., are known to be water borne. These pathogens are commonly transmitted through drinking water and cause infection of the intestinal tract. It is therefore, necessary to employ treatment facilities to purify water and to provide safe drinking water (Potable water).
Water that is free from diseases producing organism and chemical substances deleterious to health is called potable water
The main operations employed in water purification to produce potable water are: (i) sedimentation, (ii) filtration, and (iii) chlorination (fig 1). Sedimentation removes large particulate matter which settles at the bottomMost microorganisms are removed during coagulation with aluminium sulphate and sand filtration and subsequent treatment of water with chlorine (0.2 – 2 mg free chlorine per liter) will ensure its potability.
Fig. 1 Main Operation in drinking water purification

 `
`
Microbiological Quality: Water can be perfectly clear in appearance and free from odour and taste and yet, be contaminated by microorganisms. Pathogenic organisms enter into water through sewage contamination or discharges from animals or humans into the reservoirs. The coliforms (E.coli and related organisms), Streptococcus faecalis and Clostridium perfringens which are normal inhabitants of the large intestine of animals and humans enter water supplies through faecal contamination. The presence of any of these bacterial species in water is evidence of sewage or faecal pollution. Techniques are available by which the presence of these specific groups can be easily identified. The routine bacteriological examination contains consists of (i) Plate count to determine the number of bacteria present, and (ii) biochemical test to reveal the presence of coliform bacteria since these are indicator organisms for fecal contamination. Figure 2 shows a general laboratory testing scheme for detection of cliform group of bacteria in water.
A variety of other bacteria and organisms which may not be serious pathogens including faecal streptococci, slime forming bacteria, Sulphur bacteria, algae etc. may also cause problems of odour, color and taste and it is essential that be eliminated from the drinking water.
Fig. 2 Laboratory tests for detecting contamination by coliforms

Presumptive test



| Download this lecture as PDF here |
MICROBIAL SPOILAGE OF FRESH FOOD
Food is said to be spoilt if there is rotting i.e., bad smell, fermentation ie, bubbles/gas in the food or mold ie, spongy growth on the food stuff. Formation of soft spots or soft brown spots on fruits and vegetables is also food spoilage. Foods get spoilt mainly due to the presence of micro organisms, enzymes (present in foods), insects, worms, and rats.
1. Presence of micro-organisms: Micro-organisms spoil food items when the condition for their growth is ambient. Like all living beings micro-organisms require air, moisture, right temperature and food to grow and multiply. The situations which provide ambient conditions for growth of micro-organisms resulting in spoilage of foods are as follows,
- Food having high moisture content
- Air around the food containing micro organisms
- Foods kept for a long time at room temperature
- Skin of fruits and vegetables getting damaged, thus exposing the food to micro organisms.
- Foods with low salt, sugar or acid content.
Food product | Type of microorganism | Common spoilage organisms |
Fruits and vegetables | Bacteria | Erwinia, Pseudomonas, Corynebacteria (mainly vegetable pathogens; will rarely spoil fruit) |
Fungi | Aspergillus, Botrytis, Geotrichum, Rhizopus, Penicillium, Cladosporium, Alternaria, Phytophora, various yeasts | |
Fresh meat, poultry, and seafood | Bacteria | Acinetobacter, Aeromonas, Pseudomonas, Micrococcus, Achromobacter, Flavobacterium, Proteus, Salmonella |
Fungi | Cladosporium, Mucor, Rhizopus, Penicillium, Geotrichum, Sporotrichum, Candida, Torula, Rhodotorula | |
Milk | Bacteria | Streptococcus, Leuconostoc, Lactococcus, Lactobacillus, Pseudomonas, Proteus |
High sugar foods | Bacteria | Clostridium, Bacillus, Flavobacterium |
Fungi | Saccharomyces, Torula, Penicillium |
2. Presence of enzymes: Enzymes are organic catalysts found in all plants and animals. Enzymes help in ripening of fruits and vegetables. If a ripe fruit is kept for few days, it will become soft, develop black spots and will start smelling bad. This is due to continued action of enzymes.

3. Insects, worms and rats: Small insects and worms eat the food grains. They make small holes in the grain and at times convert the grain to a fine powder. The food grain thus become unfit for human consumption.

| Download this lecture as PDF here |
Principles of Food Preservation
A good method of food preservation is one that slows down or prevents altogether the action of the agents of spoilage. Also, during the process of food preservation, the food should not be damaged. In order to achieve this, certain basic methods were applied on different types of foods. For example in earlier days, in very cold weather condition, ice was used to preserve foods. Thus, very low temperature became an efficient method for preventing food spoilage. Let us now list the principles of food preservation.
1. Removal of micro-organisms or inactivating them: This is done by removing air, water (moisture), lowering or increasing temperature, increasing the concentration of salt or sugar or acid in foods. If you want to preserve green leafy vegetables, you have to remove the water from the leaves so that micro organisms cannot survive. You do this by drying the green leaves till all the moisture evaporates.
2. Inactivating enzymes: Enzymes found in foods can be inactivated by changing their conditions such as temperature and moisture, when you preserve peas, one of the methods of preservations is to put them for a few minutes in boiling water. This method also known as blanching inactivates enzymes and thus, helps in preserving the food.
3. Removal of insects, worms and rats: By storing foods in dry, air tight containers the insects, worms or rats are prevented from destroying it.
Control
Control of microorganisms
- Heat
- Cold
- Drying
- Acids
- Sugar and salt
- Oxygen concentration
- Smoke
- Radiation
- Chemicals (preservatives)
Control of enzymes
- Heat
- Oxygen removal
- Acids
- Chemicals (antioxidants)
Control of Other factors
- Protective packaging
- Sanitation
Preservation methods
1. Thermal processing
Application of heat
- Inactivate enzymes
- Kill microorganisms. Most bacteria are killed in the range 82-93°c. Spores are not destroyed even by boiling water at 100°c for 30 min.
- To ensure sterility (total microbial destruction, including spores), a temperature of 121°c must be maintained for 15 min or longer.
Various methods are –
- Blanching

- Pasteurization
- Sterilization
- Boiling
- Steam under pressure
2. Removal of heat (cold processing)
- Lowering temperature of food
- Decreases the rate of enzymatic, chemical and microbial reactions in food
- Storage life is extended
Various methods are –
- Refrigeration
- Freezing
3. Control of water content (drying)
- Microorganisms require free water
- Free water is removed from the food and therefore, is unavailable to microbial cells
- Multiplication will stop
- Water unavailable for chemical/biochemical reactions
- Storage life extended
Various methods are –
- Freezing
- Physical removal of water from food (dehydration)
- Removal of some of the water from food (concentration)
- Addition of substances that bind water in food, making it unavailable (sugar, salts)
4. Radiation
- Ionizing radiation
- Inactivate microorganisms in food
- Destroy storage pests
- Inactivate enzymes
Various methods are –
- Infrared radiation
- Ultraviolet radiation
5. Atmosphere composition
- Removal of oxygen
- Inhibits o2-dependant enzymatic and chemical reactions
- Inhibits growth of aerobic microorganisms
Various methods are –
- Paraffin wax
- Nitrogen backflushed bags (potato chips)
- Controlled atmosphere storage
- Vacuum packaging of fresh food (cured meats)
6. Fermentation
- Specific microorganisms are used (starter cultures)
- Facilitate desirable chemical changes
- Longer storage life
- Produce acids, alcohol that will prevent growth of undesirable microorganisms
- Produce antimicrobial substances
- Addition of chemicals
Various chemicals used are –
- Acids (inhibit microbial growth and enzymatic reactions)
- Organic acids (acetic, citric, tartaric acids)
- Inorganic acids (hydrochloric, phosphoric acids)
- Food grade, comply w/regulations
- Antioxidants (to delay oxidative rancidity)
- Antimicrobial agents:
- sodium propionate (mould inhibitor)
- sodium benzoate (antibacterial)
- sugar and salt (high concentrations)
8. Smoke
- Contains preservative chemicals (eg. formaldehyde) from the burning wood
- Heat also helps destroy microorganisms
- Heat dries the food
9. Curing (Salt and Sugar)
- Salt binds with water molecules and thus acts as a dehydrating agent in foods.
- Impair the conditions under which pathogens cannot survive.
- Curing is used with certain fruits and vegetables. ( sauerkraut, pickles),
- Meats can be submerged in a salt solution known as brine
PRESERVATION BY USING CHEMICALS
A preservative is defined as only substance which is capable of inhibiting, retarding or arresting the growth of microorganisms.
Microbial spoilage of food products is also controlled by using chemical preservatives. The inhibitory action of preservatives is due to their interfering with the mechanism of cell division, permeability of cell membrane and activity of enzymes.
Pasteurized squashes, cordials and crushes have a cooked flavour. After the container is opened, they ferment and spoil within a short period, particularly in a tropical climate. To avoid this, it is necessary to use chemical preservatives. Chemically preserved squashes and crushes can be kept for a fairly long time even after opening the seal of the bottle. It is however, essential that the use of chemicals is properly controlled, as their indiscriminate use is likely to be harmful. The preservative used should not be injurious to health and should be non-irritant. It should be easy to detect and estimate.
Two important chemical preservatives are permitted to beverages according to the FPO (1955).
1. Sulphur dioxide and
2. Benzoic acid
SULPHUR DIOXIDE
It is widely used throughout the world in the preservation of juice, pulp, nectar, squash, crush, cordial and other products. It has good preserving action against bacteria and moulds and inhibits enzymes, etc. In addition, it acts as an antioxidant and bleaching agent. These properties help in the retention of ascorbic acid, carotene and other oxidizable compounds. It also retards the development of nonenzymatic browning or discolouration of the product. It is generally used in the form of its salts such as sulphite, bisulphate and metabisulphite.
Potassium metabisulphite (K2O 2So2 (or) K2S2O5) is commonly used as a stable source of So2. Being a solid, it is easier to use than liquid (or) gaseous So2.It is fairly stable in neutral (or) alkaline media but decomposed by weak acids like carbonic, citric, tartaric acid and malic acids. When added to fruit juice (or) squash it reacts with the acid in the juice forming the potassium salt and So2, which is liberated and forms sulphurous acid with the water of the juice. The reactions involved are as follows
Potassium Potassium Sulphur
Meta bisulphate + Citric acid ® Citrate + dioxide + H2O
SO2 + H2O ® H2SO3 (Sulphurous acid)
SO2 has a better preservative action than sodium benzoate against bacteria and moulds. It also retards the development of yeasts in juice, but cannot arrest their multiplication, once their number has reached a high value.
It is well known that fruit juices with high acidity do not undergo fermentation readily. The preservative action of the fruit acid its due to is hydrogen ion concentration. The pH for the growth of moulds ranges from 1.5 to 8.5, that of yeasts from 2.5-8.0, and of bacteria from 4.0 to 7.5.As fruit beverage like citrus squashes and cordials have generally a pH of 2.5 to 3.5, the growth of moulds and yeasts in them cannot be prevented by acidity alone. Bacteria, however, cannot grow. The pH is therefore, of great importance in the preservation of food product and by regulating it, one or more kinds of microorganisms in the beverage can be eliminated.
The concentration of So2 required preventing the growth of mirgroorganism at different pH levels are as under.
pH | S.ellipsoideus | Mucor | Penicillium | Mixed bacteria |
2.5 | 200 | 200 | 300 | 100 |
3.5 | 800 | 600 | 600 | 300 |
7.0 | Above 5000 | Above 5000 | Above 5000 | Above 1000 |
The toxicity of So2 increases at high temperature. Hence its effectiveness depends on the acidity, pH, temperature and substances present in fruit juice.
According to FPO, the maximum amount of So2 allowed in fruit juice is 700 ppm, in squash, crush and cordial 350 ppm and in RTS and nectar 100 ppm. The advantages of using So2 are a) It has a better preserving action than sodium benzoate against bacterial fermentation b) it helps to retain the colour of the beverage for a longer time than sodium benzoate ( c) being a gas, it helps in preserving the surface layer of juices also (d) being highly soluble in juices and squashes, it ensures better mixing and hence their preservation and (e) any excess of So2 present can be removed either by heating the juice to about 71oC or by passing air through it or by subjecting the juice to vacuum. This causes some loss of the flavouring materials due to volatilization, which can be compensated by adding flavours.
Disadvantages (or) limitations
- It cannot be used in the case of some naturally coloured juices like those of jamun, pomegranate, strawberry, coloured grapes, plum etc. on account of its bleaching action.
- It cannot also be used for juices which are to be packed in tin containers because it not only corrodes the tin causing pinholes, but also forms H2S which has a disagreeable smell and reacts with the iron of the tin container to form a black compound, both of which are highly undesirable and
- So2 gives a slight taste and colour to freshly prepared beverages but these are not serious defects if the beverage is diluted before drinking.
II. Benzoic acid
It is only partially soluble in H2O hence its salt, sodium benzoate is used. One part of sodium benzoate is soluble in 1.8 parts of water at ordinary temperature, whereas only 0.34 parts of benzoic acid is soluble in 100 parts of water. Sodium benzoate is thus nearly 170 times as soluble as benzoic acid, pure sodium benzoate is tasteless and odourless.
The antibacterial action of benzoic acid is increased in the presence of Co2 and acid e.g. Bacillus subtilis cannot survive in benzoic acid solution in the presence of Co2. Benzoic acid is more effective against yeasts than against moulds. It does not stop lactic acid and acetic acid fermentation.
The quantity of benzoic acid required depends on the nature of the product to be preserved, particularly its acidity. In case of juices having a pH of 3.5-4.0, which is the range of a majority of fruit juices, addition of 0.06 to 0.10% of sodium benzoate has been found to be sufficient. In case of less acid juices such as grape juice atleast 0.3% is necessary. The action of benzoic acid is reduced considerably at pH 5.0. Sodium benzoate is excess of 0.1% may produce a disagreeable burning taste. According to FPO its permitted level in RTS and nectar is 100 ppm and in squash, crush and cordial 600 ppm.
In the long run benzoic acid may darken the product. It is, therefore, mostly used in coloured products of tomato, jamun, pomegranate, plum, watermelon, strawberry, coloured grapes etc.
Preservation by Using Radiation
Radiation may be defined as the emission and propagation of energy through space or through a material medium. The type of radiation of primary interest in food preservation is electromagnetic.
Initially, the destruction of microorganisms in foods by ionizing radiation was referred to by terminology brought over from heat and chemical destruction of microorganisms. Although microorganisms can indeed be destroyed by chemicals, heat, and radiation, there is, nevertheless, a lack of precision in the use of this terminology for radiation-treated foods. Consequently, in 1964 an international group of microbiologists suggested the following terminology for radiation treatment of foods.24
Radappertization
Is equivalent to radiation sterilization or “commercial sterility,” as it is understood in the canning industry. Typical levels of irradiation are 3(MK) kGy.
Radicidation
Is equivalent to pasteurization— of milk, for example. Specifically, it refers to the reduction of the number of viable specific nonspore-
forming pathogens, other than viruses, so that none is detectable by any standard method. Typical levels to achieve this process are 2.5-10 kGy.
Radurization
May be considered equivalent to pasteurization. It refers to the enhancement of the keeping quality of a food by causing substantial reduction in the numbers of viable specific spoilage microbes by radiation. Common dose levels are 0.75-2.5 kGy for fresh meats, poultry, seafood, fruits, vegetables, and cereal grains.
Radappertization
Radappertization of any foods may be achieved by application of the proper dose of radiation under the proper conditions.
Preservation by Using High temperature
The use of high temperatures to preserve food is based on their destructive effects on microorganisms.
By high temperatures are meant any and all temperatures above ambient. With respect to food preservation, there are two temperature categories in common use: pasteurization and sterilization.
Pasteurization: by use of heat implies either the destruction of all disease-producing organisms (for example, pasteurization of milk) or the destruction or reduction in the number of spoilage organisms in certain foods, as in the pasteurization of vinegar. The pasteurization of milk is achieved by heating as follows:
145°F (63°C) for 30 minutes (low temperature, long time [LTLT]) 1610F (72°C) for 15 seconds (primary high temperature, short time [HTST] method) 191°F(89°C) for 1.0 second, 194°F (900C) for 0.5 second, 201°F(94°C)for0.1 second, 212°F (1000C) for 0.01 second. These treatments are equivalent and are sufficient to destroy the most heat resistant of the nonspore- forming pathogenic organisms—Mycobacterium tuberculosis and Coxiella burnetii. When six different strains of M. paratuberculosis were added to milk at levels from 40 to 100,000 colony-forming units (cfu)/mL followed by pasteurization by LTLT or HTST, no survivors survivors were detected on suitable culture media incubated for 4 months. Milk pasteurization temperatures are sufficient to destroy, in addition, all yeasts, molds, gram negative bacteria, and many gram positives. The two groups of organisms that survive milk pasteurization are placed into one of two groups: thermodurics and thermophiles. Thermoduric organisms are those that can survive exposure to relatively high temperatures but do not necessarily grow at these temperatures. The nonsporeforming organisms that survive milk pasteurization generally belong to the genera Streptococcus and Lactobacillus, and sometimes to other genera. Thermophilic organisms are those that not only survive relatively high temperatures but require high temperatures for their growth and metabolic activities. The genera Bacillus and Clostridium contain the thermophiles of greatest importance in foods. Pasteurization (to destroy spoilage biota) of beers in the brewing industry is carried out usually for 8-15 minutes at 600C.
Sterilization: means the destruction of all viable organisms as may be measured by an appropriate plating or enumerating technique. Canned foods are sometimes called “commercially sterile” to indicate that no viable organisms can be detected by the usual cultural methods employed or that the number of survivors is so low as to be of no significance under the conditions of canning and storage. Also, microorganisms may be present in canned foods that cannot grow in the product by reason of undesirable pH, oxidation-reduction potential (Eh), or temperature of storage.
Preservation by Using Low temperature
The use of low temperatures to preserve foods is based on the fact that the activities of food borne microorganisms can be slowed at temperatures above freezing and generally stopped at subfreezing temperatures. The reason is that all metabolic reactions of microorganisms are enzyme catalyzed and that the rate of enzyme catalyzed reactions is dependent on temperature.
With a rise in temperature, there is an increase in reaction rate. The temperature coefficient (Q10) may be generally defined as follows:
![]()
The Qi0 for most biological systems is 1.5-2.5, so that for each 100C rise in temperature within the suitable range, there is a twofold increase in the rate of reaction. For every 100C decrease in temperature, the reverse is true.
The term psychrophile was coined by Schmidt- Nielsen in 1902 for microorganisms that grow at O0C.30 This term is now applied to organisms that grow over the range of subzero to 200C, with an optimum range of 10-150C.44 Around 1960,
the term psychrotroph (psychros, cold, and trephein, to nourish or to develop) was suggested for organisms able to grow at 5°C or below.1147 It is now widely accepted among food microbiologists that a psychrotroph is an organism that can grow at temperatures between 00C and 7°C and produce visible colonies (or turbidity) within 7-10 days. Because some psychrotrophs can grow at temperatures at least as high as 430C, they are, in fact, mesophiles. By these definitions, psychrophiles would be expected to occur only on products from oceanic waters or from extremely cold climes. The organisms that cause the spoilage of meats, poultry, and vegetables in the 0-50C range would be expected to be
psychrotrophs.
Methods of freezing
There are various methods of freezing
1. Sharp Freezing (Slow freezing)
This technique, first used in 1861, involves freezing by circulation of air, either naturally or with the aid of fans. The temperature may vary from –15 to –29oC and freezing may take from 3 to 72 hours. The ice crystals formed one large and rupture the cells. The thawed tissue cannot regain its original water content. The first products to be sharp frozen were meat and butter. Now-a-days freezer rooms are maintained at –23 to –29oC or even lower, in contrast to the earlier temperature of –18oC.
2. Quick freezing
In this process the food attains the temperature of maximum ice crystal formation (0 to – 4oC) in 30 min or less. Such a speed results in formation of very small ice crystals and hence minimum disturbance of cell structure. Most foods are quick frozen by one of the following three methods:
a) By direct immersion
Since liquids are good heat conductors food can be frozen rapidly by direct immersion in a liquid such as brine or sugar solution at low temperature. Berries in sugar solution packed fruit juices and concentrates are frozen in this manner. The refrigeration medium must be edible and capable of remaining unfrozen at –18oC and slightly below. Direct immersion equipments such as ottenson Brine freezer, Zarotschenzeff ‘Fog’ freezer, T.V.A. freezer, Bartlett freezer etc. of commercial importance earlier are not used today.
Advantages
- There is perfect contact between the refrigerating medium and the product, hence the rate of heat transfer is very high.
- Fruits are frozen with a coating of syrup which preserves the colour and flavour during storage.
- The frozen product is not a solid block because each piece is separate.
Disadvantages
- Brine is a good refrigerating medium but it cannot be used for fruits.
- It is difficult to make a syrup that will not become viscous at low temperature.
- The refrigeration temperature must be carefully controlled, as at high temperature the medium will enter the product by osmosis and at low temperature the medium may freeze solid.
- It is very difficult to maintain the medium at a definite concentration and also to keep it free from dirt and contamination.
b) By indirect contact with refrigerant
Indirect freeing may be defined as freezing by contact of the product with a metal surface which is itself cooled by freezing brine or other refrigerating media. This is an old method of freezing in which the food or package is kept in contact with the passage through the refrigerant at –18 to -46oC flows. Knowles Automatic Package feezer, Patterson continuous plate freezer, FMC continuous can freezer and Birds eye freezers are based on this principle.
c) By air blast
In this method, refrigerated air at –18 to –34oC is blown across the material to be frozen. The advantages claimed for quick freezing over slow freezing (sharp freezing) are (1) smaller (size) ice crystals are formed, hence there is less mechanical destruction of intact cells of the food (2) period for ice formation is shorter, therefore, there is less time for diffusion of soluble material and for separation of ice (3) more rapid preservation of microbial growth and (4) more rapid slowing down of enzyme action.
3) Cryogenic freezing
Although most foods retain their quality when quick frozen by the above methods, a few require ultrafast freezing. Such materials are subjected to cryogenic freezing which is defined as freezing at very low temperature (below –60oC). The refrigerant used at present in cryogenic freeing are liquid nitrogen and liquid CO2. In the former case, freezing may be achieved by immersion in the liquid, spraying of liquid or circulation of its vapour over the product to be frozen.
4. Dehydro-freezing
This is a process where freezing is proceded by partial dehydration. In case of some fruits and vegetables about 50% of the moisture is removed by dehydration prior to freezing. This has been found to improve the quality of the food. Dehydration does not cause deterioration and dehydro frozen foods are relatively more stable.
5. Freeze drying
In this process food is first frozen at –18oC on trays in the lower chamber of a freeze drier and the frozen material dried (initially at 30oC for 24 hrs and then at 20oC). Under high vacuum (0.1 mm Hg) in the upper chamber. Direct sublimation of the ice takes place without passing through the intermediate liquid stage. The product is highly hygroscopic, excellent in taste and flavour and can be reconstituted readily. Mango pulp, orange juice concentrate, passion fruit juice and guava pulp are dehyderated by this method.
| Download this lecture as PDF here |
What is Fermentation ? Fermentation is the chemical transformation of organic substances into simpler compounds by the action of enzymes, complex organic catalysts, which are produced by microorganisms such as molds, yeasts, or bacteria. Enzymes act by hydrolysis, a process of breaking down or predigesting complex organic molecules to form smaller (and in the case of foods, more easily digestible) compounds and nutrients. For example, the enzyme protease breaks down huge protein molecules first into polypeptides and peptides, then into numerous amino acids, which are readily assimilated by the body. The enzyme amylase works on carbohydrates, reducing starches and complex sugars to simple sugars. And the enzyme lipase hydrolyzes complex fat molecules into simpler free fatty acids. These are but three of the more important enzymes. There are thousands more, both inside and outside of our bodies. In some fermentations, important by-products such as alcohol or various gases are also produced. The word “fermentation” is derived from the Latin meaning “to boil,” since the bubbling and foaming of early fermenting beverages seemed closely akin to boiling.
Fermented foods often have numerous advantages over the raw materials from which they are made. As applied to soyfoods, fermentation not only makes the end product more digestible, it can also create improved (in many cases meatlike) flavor and texture, appearance and aroma, synthesize vitamins, destroy or mask undesirable or beany flavors, reduce or eliminate carbohydrates believed to cause flatulence, decrease the required cooking time, and increase storage life. Most fermentations are activated by either molds, yeasts, or bacteria, working singularly or together. The great majority of these microorganisms come from a relatively small number of genera; roughly eight genera of molds, five of yeasts, and six of bacteria. An even smaller number are used to make fermented soyfoods: the molds are Aspergillus , Rhizopus , Mucor , Actinomucor , and Neurospora species; the yeasts are Saccharomyces species; and the bacteria are Bacillus and Pediococcus species plus any or all of the species used to make fermented milk products. Molds and yeasts belong to the fungus kingdom, the study of which is called mycology. Fungi are as distinct from true plants as they are from animals. The study of all microorganisms is called microbiology. While microorganisms are the most intimate friends of the food industry, they are also its ceaseless adversaries. They have long been used to make foods and beverages, yet they can also cause them to spoil. When used wisely and creatively, however, microorganisms are an unexploitable working class, whose very nature is to labor tirelessly day and night, never striking or complaining, ceaselessly providing human beings with new foods. Like human beings, but unlike plants, microorganisms cannot make carbohydrates from carbon dioxide, water, and sunlight. They need a substrate to feed and grow on. The fermented foods they make are created incidentally as they live and grow.
Human beings are known to have made fermented foods since Neolithic times. The earliest types were beer, wine, and leavened bread (made primarily by yeasts) and cheeses (made by bacteria and molds). These were soon followed by East Asian fermented foods, yogurt and other fermented milk products, pickles, sauerkraut, vinegar (soured wine), butter, and a host of traditional alcoholic beverages. More recently molds have been used in industrial fermentation to make vitamins B-2 (riboflavin) and B-12, textured protein products (from Fusarium and Rhizopus in Europe) antibiotics (such as penicillin), citric acid, and gluconic acid. Bacteria are now used to make the amino acids lysine and glutamic acid. Single-celled protein foods such as nutritional yeast and microalgae (spirulina, chlorella) are also made in modern industrial fermentations.
For early societies, the transformation of basic food materials into fermented foods was a mystery and a miracle, for they had no idea what caused the usually sudden, dramatic, and welcomed transformation. Some societies attributed this to divine intervention; the Egyptians praised Osiris for the brewing of beer and the Greeks established Bacchus as the god of wine. Likewise, at many early Japanese miso and shoyu breweries, a small shrine occupied a central place and was bowed to daily. In ancient times fermentation joined smoking, drying, and freezing as basic and widely practiced food preservation techniques. Wang and Hesseltine (1979) note that “Probably the first fermentation were discovered accidentally when salt was incorporated with the food material, and the salt selected certain harmless microorganisms that fermented the product to give a nutritious and acceptable food.” The process was taken a step further by the early Chinese who first inoculated with the basic foods with molds, which created enzymes; in salt-fermented soyfoods such as miso, soy sauce, soy nuggets, and fermented tofu, these aided salt-tolerant yeasts and bacteria??.
A Brief History of Fermentation in the West. The origins of microbiology (other than the general knowledge of fermented foods which existed worldwide since ancient times) can be traced back to the invention of the compound microscope in the late 1500s. This relatively simple tool soon revolutionized man’s knowledge of the heretofore invisible microbial world. In 1675 the Dutch merchant Anton van Leeuwenhoek, the greatest of the early microscopists, saw and reported one-celled organisms, which he called “animacules.” (Today they are called “protozoa.”) The discovery electrified the scientific world of the time. Then in 1680, using a microscope that magnified the diameter of each object 300-fold, he looked at yeast and found them to consist of tiny spheroids. While the protozoa were clearly alive, the yeast did not appear to be. No connection was drawn between the existence of these tiny organisms and the well known phenomenon of fermentation. So for 150 years after van Leeuwenhoek’s pioneering observations, it was hardly thought that these minute organisms could be important enough to deserve serious study.
The early 1800s saw a great increase of interest in microbiology in Europe. The scientific period began with great advances in botany, increased interest in microscopy, and willingness to investigate individual organisms. The two major problems that would challenge the greatest researchers in the new field of microbiology concerned the basic nature of the fermentation process and the basic nature of enzymes. The scientific breakthroughs that would lead to the unraveling of the mysteries of fermentation starting in the 1830s were made primarily by French and German chemists. In the late 1700s Lavoisier showed that in the process of transforming sugar to alcohol and carbon dioxide (as in wine), the weight of the former that was consumed in the process equaled the weight of the latter produced. The first solid evidence of the living nature of yeast appeared between 1837 and 1838 when three publications appeared by C. Cagniard de la Tour, T. Swann, and F. Kuetzing, each of whom independently concluded as a result of microscopic investigations that yeast was a living organism that reproduced by budding. The word “yeast,” it should be noted, traces its origins back to the Sanskrit word meaning “boiling.” It was perhaps because wine, beer, and bread were each basic foods in Europe, that most of the early studies on fermentation were done on yeasts, with which they were made.
The view that fermentation was a process initiated by living organisms soon aroused fierce criticism from the finest chemists of the day, especially Justus von Liebig, J.J. Berzelius, and Friedrich Woehler. This view seemed to give new life to the waning mystical philosophy of vitalism, which they had worked so hard to defeat. Proponents of vitalism held that the functions of living organisms were due to a vital principal distinct from physico-chemical forces, that the processes of life were not explicable by the laws of physics and chemistry alone, and that life was in some part self determining. As we shall soon see, the vitalists played a key role in debate on the nature of fermentation. A long battle ensued, and while it was gradually recognized that yeast was a living organism, its exact function in fermentations remained a matter of controversy. The chemists still maintained that fermentation was due to catalytic action or molecular vibrations.
The debate was finally brought to an end by the great French chemist Louis Pasteur (1822-1895) who, during the 1850s and 1860s, in a series of classic investigations, proved conclusively that fermentation was initiated by living organisms. In 1857 Pasteur showed that lactic acid fermentation is caused by living organisms. In 1860 he demonstrated that bacteria cause souring in milk, a process formerly thought to be merely a chemical change, and his work in identifying the role of microorganisms in food spoilage led to the process of pasteurization. In 1877, working to improve the French brewing industry, Pasteur published his famous paper on fermentation, Etudes sur la Biere , which was translated into English in 1879 as Studies on Fermentation . He defined fermentation (incorrectly) as “Life without air,” but correctly showed specific types of microorganisms cause specific types of fermentations and specific end products. In 1877 the era of modern medical bacteriology began when Koch (a German physician; 1843-1910) and Pasteur showed that the anthrax bacillus caused the infectious disease anthrax. This epic discovery led in 1880 to Pasteur’s general germ theory of infectious disease, which postulated for the first time that each such disease was caused by a specific microorganism. Koch also made the very significant discovery of a method for isolating microorganisms in pure culture.
Interestingly, until his death in 1873, the eminent German chemist J. von Liebig continued to attack Pasteur’s work on fermentation, putrefaction, and infectious diseases. He recognized the similarity of these phenomena but refused to believe that living organisms were the main causative agents. Fermentation, he felt, was primarily a chemical rather than a biological process. History has shown, with the discovery of enzymes, that Pasteur was not entirely right, nor Liebig entirely wrong.
The work of Pasteur and his many colleagues and predecessors opened up vast new vistas in the fields of biochemistry, microbiology, and fermentation. The term “biochemistry” was first used in English in 1869, but this new science of the application of chemistry to biology was generally called “physiological chemistry” until the early 1900s. The two outstanding pioneers were Liebig and Pasteur. The term “microbiology” was first used in English in 1885, long after Pasteur’s major discoveries. But basic knowledge of this new science of the study of minute living organisms closely related to human activity or welfare did not begin to enter the popular consciousness until the early 1900s. At about this time the scientific breakthroughs of the 1870s and 1880s had begun to produce a change in people’s conception of the world around them so sweeping and profound as to be termed revolutionary. Food microbiology was finally set on a scientific foundation, based on the action of specific microorganisms. A rational theory of infectious diseases (which formerly were not differentiated from one another) set people’s minds free from the age-old fear of vengeance from an unknowable and invisible disease-causing entity. And the ancient theory of spontaneous generation of lower life forms, which said they could arise de novo and fully formed from decomposing matter, was replaced by the verifiable theory of biogenesis. For the first time people began to accept the fact that they shared their environment with multitudes of minute organisms that exerted an ongoing powerful influence on human life. This new world view, among other things, provided a tremendous stimulus for new research on fermented foods.
Although showing that fermentation was generally the result of the action of living microorganisms was an epic breakthrough, it did not explain the basic nature of the fermentation process, or prove that it was caused by the microorganisms that were apparently always present. As early as the late 1700s it had been recognized that there was another type of chemical change that resembled the yeast fermentation in some respects. This was the sort of changes that occur, for example, in the digestion of food. In 1752 Reamur, in studying the digestive processes of a falcon, showed that its digestive juices were able to dissolve meat. In 1785 William Irvine discovered that aqueous extracts of sprouted barley caused liquefaction of starch. The first clear recognition of what were later called “enzymes” came in 1833 when two French chemists, A. Payen and J.F. Persoz, made a more detailed investigation of the process of solubilizing starch with a malt extract to form a sugar that they called “maltose.” They called the agent responsible for this transformation “diastase” and they showed that it was destroyed or inactivated by boiling, that without undergoing permanent change itself, a small amount of diastase could convert a large amount of starch to sugar, and that it could be concentrated and purified by precipitation with alcohol. In 1835 the German naturalist Swann, mentioned above for his early work with fermentation, isolated a substance from gastric juice which could bring about the dissolution of meat but which was not an acid. He called it “pepsin” from a Greek word meaning “digestion.” It soon became fashionable to call organic catalysts such as diastase and pepsin “ferments,” because digestion and fermentation, both allied with life, seemed to be somewhat similar processes. Under the influence of the vitalists, ferments were grouped into two types: those involved with life process were called “organized ferments” and those which were not (like pepsin) were merely “unorganized ferments.” A relation between the two types of ferments was suspected by many, and in 1858 M. Traube put forward the theory that all fermentations were due to ferments, definite chemical substances he regarded as related to the proteins and produced in the cells by the organism. In 1876, to reduce confusion that existed concerning the two types of ferments, the German physiologist Wilhelm Kuehne suggested that an unorganized ferment, acting in the absence of life, be called an “enzym,” after the Greek words meaning “in yeast;’ in 1881 this term was anglicized to “enzyme” by William Roberts, and it had begun to catch on by the 1890s.
Many scientists, including Pasteur, had attempted unsuccessfully to extract the fermentation enzyme from yeast. Success came finally in 1897 when the German chemist Eduard Buechner ground up yeast, extracted a juice from them, then found to his amazement that this “dead” liquid would ferment a sugar solution, forming carbon dioxide and alcohol . . . just like living yeasts. Clearly the so-called “unorganized ferments” behaved just the organized ones. From that time on the term “enzyme” came to be applied to all ferments. The term “ferment” dropped out of the scientific vocabulary altogether and the vitalist position collapsed, never to recover. Thereafter it was agreed that only one set of laws applied to all things, both animate and inanimate, and that there was no special vital force which characterized living things and acted under different laws. And it was finally understood that fermentation is caused by enzymes which are produced by microorganisms. In 1907 Buechner won the Nobel Prize in chemistry for his work, which opened a new era in enzyme and fermentation studies.
The sciences of microbiology, biochemistry, fermentation technology, mycology, and bacteriology all shared a deep interest in the nature and working of enzymes. Yet still by the early 1900s no one knew exactly what enzymes were or how they acted. As the agricultural microbiologist Conn asked in 1901, “How can they produce chemical actions without being acted upon or entering into the reactions? Are enzymes fully lifeless or semi-living? We still do not know the fundamental mystery of fermentation.” Gradually an understanding of enzymes and catalysts developed. In 1905 Harden and Young discovered coenzymes, agents necessary for the action of enzymes. In 1926 the American biochemist J.B. Sumner first purified and crystallized an enzyme (urease) and showed that it was a protein, more precisely a protein catalyst. Eventually enzymes came to be seen as the key catalysts in all the life processes, each highly specialized in its catalytic action and generally responsible for only one small step in complex, multi-step biochemical reactions. Enzymes are still produced only by living organisms, both animals and plants; they have never? been synthesized.
Advances in microbiology and fermentation technology have continued steadily up until the present. For example, in the late 1930s it was discovered that microorganisms could be mutated with physical and chemical treatments to be higher yielding, faster growing, tolerant of less oxygen, and able to use a more concentrated medium. Strain selection and hybridization developed as well, affecting most modern food fermentations (Hesseltine and Wang 1977).
A Brief History of Fermentation in East Asia . Traditional fermented foods play an unusually extensive role in East Asia food systems. These fermented foods have a number of important distinguishing characteristics: a number of the most important fermentations use molds; dairy products and other animal proteins (excepting fish) are not widely used, as they are in the West; and modern fermentation processes and technology are based largely on traditional processes, yet are extremely advanced and sophisticated.
The main use of molds has been in the process of making koji (mold-fermented grains and/or soybeans), which serves as a source of more than 50 enzymes in a subsequent fermentation in much the same way that, in the West, the enzymes of malt (steeped and sprouted barley or other cereal grains) are used to make alcoholic beverages.
Since ancient times the koji making process has been unique to East Asia, where it has been used in the preparation of fermented foods such as miso, soy sauce, soy nuggets, sake, shochu (spirits), and rice vinegar ( yonezu ). The only traditional East Asian fermented soyfood not prepared with molds is Japan’s natto, and its relatives thua-nao in Thailand and kinema in Nepal; these are bacterial fermentations. Some have suggested that molds are widely used since they grow well in areas having a humid climate and long rainy season during the warm months. In the West mold fermented foods are limited primarily to a number of cheeses characterized by their strong flavors and aromas: Camembert, Blue, Brie, and related types. Because of the widespread use of mold-fermented foods in East Asia, the word “mold” there has a rather positive connotation, something like “yeast” in the West. Most Westerners still have a deep-seated prejudice against moldy products, and they generally associate the word “mold” with food spoilage, as in “moldy bread.”
Surprisingly little has been published in English about the history of fermentation and knowledge of the fermentation process in East Asia, especially the history prior to the 1870s and 1880s, when the new science of microbiology was introduced from the West. The few works that do exist will be cited later.
The earliest records of the koji-making process can be traced back to at least 300 BC in China and to the third century AD in Japan. Molds differ in one important respect from yeasts and bacteria in that they can be easily observed with the naked eye (without a microscope) and their growth, form, and color noted. In East Asia it was probably understood that fermentation was a life process long before it was in the West. By the sixth century AD, as recorded in the Ch’i-min yao-shu (the earliest encyclopedia of agriculture), the Chinese had distinct names for two types of molds used in fermented soyfoods; what we now call Aspergillus was then called “yellow robe” and Rhizopus was called “white robe.” These cultures were carefully distinguished and propagated from year to year. By the 10th century a koji starter or inoculum was deliberately being used in the preparation of koji for fermented foods (Tamiya 1958; Sakaguchi 1972; 1979).
From these early times until the 1870s the traditional fermented foods industries in East Asia apparently advanced largely by an empirical, trial-and-error process without the benefit of general scientific research into the nature of microorganisms and of the fermentation process, and without any general theories in these areas.
Prior to 1870, makers of East Asian fermented foods were unaware of the basic nature of the fermentation process of microorganisms, enzymes, and their respective interactions. Makers of koji had no idea what caused the grains and/or soybeans to become covered with a fragrant white mycelium after several days of incubation in a warm koji room, or what later transformed the koji almost magically into delicious, savory seasonings such as miso, shoyu, or soy nuggets, or into heady beverages such as sake.
The advances in food fermentations resulting from the exchange of people and ideas was most pronounced in Japan. The first generation of European scientists there plunged in to their investigations of the many fermented foods with great curiosity and enthusiasm. One of their first subjects of research was the koji mold, now known as Aspergillus oryzae, and the various foods in which it was used, especially sake and shoyu, which were major sources of tax revenue for the Meiji government. Tradition ascribed the introduction of sake brewing in Japan to some emigrants from Korea at about the end of the third century AD; they doubtless learned the process from China, where it had long been practiced. One of the earliest accounts of sake production by a Westerner appeared in 1874 when Dr. J.J. Hoffmann, a German professor in the medical school of today’s Tokyo University, published a translation of an article on sake from a Japanese encyclopedia of 1714. In 1884 Ferdinand J. Cohn, a Polish botanist and microbiologist, first gave the koji mold its present name, Aspergillus oryzae . After 1884 the koji mold was referred to as Aspergillus oryzae (Ahlburg) Cohn, in recognition of Ahlburg’s earliest accurate description.
Another pioneer in the field of koji research was Atkinson, who had a BS degree from London and was a professor of analytical and applied chemistry at Tokyo University. In 1878, after visiting sake factories, he wrote “On Sake Brewing,” which contained a preliminary description of the koji-making process and mentioned the word “koji.” In 1881, after extensive research with his assistant Mr. Nakazawa at the koji plant of Mr. J. Kameyama in Yushima near Tokyo, he published two major articles. In his 73-page “On the Chemistry of Sake Brewing,” he gave a detailed account of koji making in underground caves in Tokyo and an analysis of its composition.
Another early leader in the fields of microbiology and fermented soyfoods was K. Saito. He did excellent early investigations on the shoyu fermentation, named the primary tempeh mold ( Rhizopus oligosporus ) in 1905, and was an authority on yeasts and molds. Likewise K.N. Yabe did important early work in bacteriology and in natto fermentation.
Two other early pioneers in the introduction of microbiology and fermentation science to Japan were Dr. Teizo Takahashi (1875-1952) and his student Dr. Kinichiro Sakaguchi (1897-), both of whom were professors in the Department of Agricultural Chemistry of Tokyo University. Both men did numerous important studies relating to miso, shoyu, and the koji mold, Aspergillus.
During the 20th century, Japanese microbiologists have made many important contributions to the development of applied and industrial microbiology, including the manufacture of fermented soyfoods, as well summarized by Tamiya (1958) and Sakaguchi (1972). Until quite recently, their strength was more in the area of application of scientific knowledge than in pioneering basic scientific and microbiological breakthroughs. From the early 1900s, important studies on the koji mold and its enzymes were done by Japanese scientists. Important advances in enzymology, with much of the work done on koji molds, began in the 1920s. In 1928 Miyazaki developed the combined Amylo-Koji process. By the 1950s Japanese scientists had isolated various protease and amylase enzymes, induced mutations, and used them commercially. They also developed the technology for the microbial production of L-glutamic acid and monosodium glutamate ( MSG), lysine and other amino acids, flavor enhancing nucleotides such as inosinic acid, and organic acids. They used the koji mold Aspergillus oryzae in the commercial production of enzymes including proteases, amylases, amyloglucosidase, and lipase. They made microbial rennet and numerous other products. Indeed in the period following World War II, Japan became the world leader in the field of industrial fermentations. Wang and Hesseltine (1979) have suggested that this may have been “in large part due to the food fermentation base from which it launched its industrialization of micoorganisms.”
| Download this lecture as PDF here |
Microbes are an integral part of soil and contribute to soil and plant health. Microorganisms have the ability to fix atmospheric nitrogen, solubilize and mobilize phosphorus, produce antibiotics and disease suppressing molecules. Owing to these properties, they are used in agriculture as biofertilizers and biopesticides. They are also important in the treatment of solid waste and sewage. They clean up the environment by degradation of several pollutants like pesticides, hydrocarbons, dyes and paints. They also help in the enhanced recovery of oil and metals from low grade ores or aqueous streams.
Man is a host to variety of pathogenic bacteria, protozoa and viruses. They can cause various infectious and non-infectious diseases. In order to control the disease and its transmission, it is essential to isolate and identify the causal agent from blood, sputum, urine, stool or pus. Various cultural and molecular methods can be employed for identification of pathogen. Sterilization techniques, use of disinfectants and vaccination can help control transmission of disease.
Biofertilizers
Biofetilizers are the products containing living cells of different types of microorganisms that enrich the nutrient quality of soil. The main sources of biofertilizers are bacteria, fungi and cyanobacteria (blue green algae). Most biofertilizers belong to one of the following categories: nitrogen fixing, phosphate solubilizing and mobilizing, and plant growth promoting rhizobacteria. Some of the major biofertilizers and target crops are given in table 8.1. Nitrogen fixing biofertilizers fix atmospheric nitrogen into forms which are readily useable by plants. These include Rhizobium, Azospirillum, Azotobacter, blue green algae and Azolla. While Rhizobium requires symbiotic association with the root nodules of legumes to fix nitrogen, others can fix nitrogen independently. Phosphate solubilizing microorganisms secrete organic acids that enhance the uptake of phosphorus by plants by dissolving rock phosphate and tricalcium phosphate. Arbuscular mycorrhizal fungi are the most common phosphorus mobilising types that are omnipresent. A group of bacteria that enhance the growth of plant through nitrogen fixation, phosphorus solubilization or production of plant growth promoting metabolites are known as Plant Growth Promoting Rhizobacteria (PGPR). Many PGPR strains have a potential to be used as microbial inoculants to enhance crop productivity.
Major biofertilisers and target crops
Biofertiliser | Target crop |
Rhizobium | Leguminous crops |
Azotobacter | Wheat, maize, cotton, mustard and vegetables (Potato, onion, tomato, brinjal and others) |
Azospirillum | Cereal crops like wheat, maize, millets, sorghum, barley; and sugarcane. |
Blue green algae (BGA) | Rice |
Azolla | Rice |
Phosphate solubilizing microorganisms | All |
Arbuscular mycorrhiza | Nursery raised crops and orchard trees |
Plant growth promoting rhizobacteria | All |
The growth in agricultural production in the last three decades has been accompanied by a sharp increase in the use of chemical fertilizers, causing serious concern. Foremost among these concerns is the effect of excessive fertilizers on the quality of soil and ground water. The use of environmental friendly biofertilizers can cut down the use of chemical fertilizers. Biofertilizers have definite advantage over chemical fertilizers. It is economical to use biofertilizers as they are a cheap source of nutrients when compared to chemical fertilizers. Biofertilisers in addition to nitrogen and phosphorus, also provides certain growth promoting substances like hormones, vitamins, and amino acids that improves the plant health and vigour. Continuous use of chemical fertilisers adversely affects the soil structure whereas biofertilizers when applied to soil improve the soil structure. The chemical fertilizers are toxic at higher doses where as biofertilizers have no toxic effects.
Nitrogen fixing bacteria
An atmosphere around us contains nearly 78% nitrogen that is in free form and is not utilized by the plants. Plants take up nitrogen in the form of ammonia or nitrate. Relatively small amount of ammonia is produced by lightning. Some ammonia also is produced industrially by the Haber-Bosch process, using an iron-based catalyst, very high pressures and fairly high temperature. But the major conversion of N2 into ammonia by the action of enzyme nitrogenase, and thence into proteins, is achieved by microorganisms in the process called nitrogen fixation (or dinitrogen fixation).
All the nitrogen-fixing organisms are prokaryotes. There are different groups of nitrogen fixing microorganisms (diazotrophs) present in the nature. These are broadly divided into three categories, viz.,
- Symbiotic microorganism
- Asymbiotic or free living
- Associative Symbiosis
Examples of nitrogen fixing microorganisms for each category are given in table
Some examples of nitrogen fixing bacteria belonging to different categories.
| Category | Examples |
Symbiotic | Rhizobium– legume symbiosis |
Free living | |
1. Aerobic | Azotobacter |
2. Facultative | Klebsiella pneumoniae |
3. Anaerobic | Clostridium |
Associative | Azospirillum |
The list of nitogen fixing bacteria is long but here we will discuss some of the important types of biofertilisers that can be considered for agrobased industries.
RHIZOBIUM INOCULANT
The bacteria of the genera Rhizobium, Bradyrhizobium, Mesorhizobium, Sinorhizobium and Azorhizobium collectively known as rhizobia, in symbiotic association with leguminous plants reduce atmospheric nitrogen. The rhizobial colonies appear raised, wet, shining, translucent or opaque with smooth margin on yeast extract mannitol agar (YEMA) medium. The legume-rhizobia symbiosis culminates in the formation nitrogen fixing root or stem nodules. These unique structures are agronomically significant as they provide alternative to the use of energy-expensive ammonium fertilizer. Not all legumes fix nitrogen. Of the three segregate families of legumes, the capacity to form nodules appear to be absent from the majority of species of Caesalpiniaceae. All members of family Mimosaceae and Fabaceae show formation of nodules with rhizobia. It is believed that legume-Rhizobium symbiosis contributes atleast 70 million metric tons N per year. The amount of nitrogen fixed varies with the strain of Rhizobium, the plant species and environmental conditions.
 |  |
| Fig Typical growth of Rhizobium on yeast mannitol agar medium with congo red | Fig Root nodules formed by extract rhizobia on mungbean plant |
The taxonomy of root and stem nodulating bacteria is in a state of transition. The initial classification of these organisms based on plant infection into 7 cross inoculation groups has been abandoned after extensive criticism. A new system of classification was proposed by Jordan (1984) in Bergey’s Manual of Systematic Bacteriology (Table ). He separated the root nodule bacteria into two genera, Rhizobium and Bradyrhizobium, based on data on numerical taxonomy, molecular characteristics and rate of growth on laboratory media. Fast growing strains were placed under genus Rhizobium whereas slow growing strains were placed in Bradyrhizobium. Since 1984, the classification has undergone lot of changes. Three additional genera, Mesorhizobium, Sinorhizobium and Azorhizobium have been recognised and many new species have been reported so far.
Rhizobium inoculation
Legume inoculation is a significant strategy for the manipulation of rhizobial microflora and improving crop productivity and soil fertility. However, in tropical soils where there is presence of adequate native rhizobia and high levels of mineral N, legume inoculation often fails. Thus there is an urgency to identify conditions where inoculation is needed. Different diagnostic measures to decide about inoculation have been suggested by various workers. Inoculation should be carried out if;
- population density of species-specific rhizobia is low,
- the same or symbiotically related legume is not grown in the area in the immediate past history
- waste-lands have to be reclaimed
- legume follows a non leguminous crop in a rotation
- soil is poor in mineral N (nitrate)
- soils are acidic, alkaline and saline.
Selection of rhizobial strains for inoculant production
A large-scale screening should be carried out to identify ideal inoculant strain for different legume crops. The criterion for selection may vary for particular soil types like acidic, sodic, saline, nitrate-rich or heavy metal contaminated. Following are some of the desirable characters for a strain to be fit for use in commercial inoculants:
1. Ability to form nodules and fix N on the target legume
2. Ability to compete in nodule formation with populations of native rhizobia present in the soil.
3. Ability to fix N across a range of environmental conditions;
4. Ability to grow well in artificial media, in inoculant carrier and in the soil
5. Ability to persist in soil, particularly for annually regenerating legumes
6. Ability to migrate from the initial site of inoculation
7. Ability to colonize the soil in the absence of a legume host
8. Ability to tolerate environmental stresses;
9. Ability to fix N with a wide range of host genotypes;
10. Genetic stability
11. Compatibility with agrochemicals.
Inoculant production
- Propagation
Rhizobia are not very particular in their nutritional requirements. Yeast-extract mannitol (YEM) medium is commonly employed for culturing of rhizobia. For commercial production of cultures, cheaper sources like sucrose, molasses and corn steep liquor can be used.
Mass scale propagation of rhizobia can be carried out using system of rotary shaker or fermentor. In shake flask culture, broth is raised in flasks with agitation by circular motion of rotary shaker. Fermentors are used for industrial scale production of bio-fertilizers. Culture vessels ranging from 5 to 1000 L can be used depending upon the requirement. The amount of inoculum culture to be added into the fermenter vessel depends on the size of the fermentors, but the ratio between the inoculum and the medium in the vessel should be maintained at 1:20 (5% inoculum rate). The broth is continuously aerated by forcing sterile air through porous stainless steel sparger. Various fermentation requirements like aeration, agitation and fermentation time vary from strain to strain. Table gives the optimum fermentation conditions for mass multiplication of rhizobial strains.
When the number of rhizobia in the broth has attained the required standard (108-109 cells ml-1) the broth should be added to the carrier for preparation of carrier-based inoculant.
Optimum fermentation conditions for mass multiplication of Rhizobium strains
________________________________________________________________
1. Type of reactor Stirred tank
2. Type of operation Batch
3. Carbon source Sucrose or malasses (3-5 g L-1)
4. Nitrogen source Corn steep liquor or yeast extract
5. pH 7.8 (controlled)
6. Temperature 280C
7. Inoculum rate 10% (V/V)
8. Inoculum count 109 cells mL-1
9. Antifoam PPG
____________________________________________________________
- Carriers for rhizobial inoculants
The medium in which rhizobia are allowed to multiply is an important factor in rhizobial culture preparation. The term ‘carrier’ is generally used for a medium that carries the live microorganisms. As per BIS specification, the carrier should be in powder form and capable of passing through 150-212 micron (72-100 mesh) IS sieve. A good carrier material should
- have high water holding capacity
- be non-toxic to rhizobia
- be easy to sterilize by autoclaving or gamma irradiation
- be readily and inexpensively available
- provide good adhesion to seed
- have pH buffering capacity
- have cation and/or anion exchange capacity.
In India, different carrier materials like peat, liginite, charcoal, rice husk, pressmud, vermiculite, soil and coir dust has been employed. Although peat is the favoured base for inoculants world over, in India high quality peat is not available. A mixture of charcoal and soil in ratio of 3:1 is most commonly used as a carrier material. The preparation of charcoal based carrier is given below.
- The carrier material is sun dried up to a moisture level of 5%. The material is ground to a desired fineness preferably to pass 100-200 mesh sieves.
- The carriers are mixed with finely powdered calcium carbonate to neutralize if they are acidic. To make charcoal more suitable for the multiplication of rhizobia, CaCO3 @ 1%, KHPO4 @ 0.5% and soil @ 25% are mixed thoroughly with it. Finally the carrier is mixed with 10% water before sterilization.
- The pretreated carrier is sterilized in an autoclave at 15 lb psi for 3-4 hr continuously.
- Broth culture of Rhizobium containing 109 cells mL-1 is added to one-third of the water holding capacity of the carrier.
- Curing
In manufacturing inoculants, a period of “curing” (maturation) after addition of broth culture to carrier improves the quality of the product. After mixing the carrier with the broth culture raw-blended carrier is kept for 24 hours for curing. During this time the rhizobia get acclimatized with the carrier.
d) Packing
After curing, the inoculant is packed in polyethylene bags (high density; 0.075 – 0.090 mm) or polypropylene bags. The packing material should have the following properties:
- should be stable towards gamma irradiation
- should be autoclavable
- should have high gas exchange capacity
- should not allow high rates of moisture loss
e) Incubation and storage
Inoculants must be incubated for a week in a room with an ambient temperature ranging from 25-30oC. During this period the bacterium multiplies and reaches to a required standard. The packets may then be stored in a cold room (4o-15oC) till its use.
Inoculant quality control
The quality of rhizobial inoculants is of great importance in ensuring field performance as well as for the commercial prospects of inoculant industry. Basically, quality means the presence of the right type of micro-organism in active form and desired numbers. Evaluation of inoculant quality by enumeration of viable rhizobia is an accurate index of inoculating potential. Numerical considerations are of such significance in determining quality of inoculant products and their success in field that the necessity for quality control systems has been recognized in various countries. In India, Bureau of Indian Standards (BIS) (formerly ISI) listed the Indian standard specifications for Rhizobium inoculants in 1977 (IS: 8268-1976). This was revised in 1986 (ISI 1986). These specifications are given in Table.
Indian Standard specifications for Rhizobium
________________________________________________________________________
Parameters Specifications
________________________________________________________________________
1. Base Carrier based
2. Cell number at the time of manufacture 108 g-1 carrier
3. Cell number at the time of expiry 107 g-1 carrier within 15 days before expiry date
4. Expiry period 6 months from the date of manufacture
5. Permissible contamination No contamination at 108 dilution
6. pH 6.0- 7.0
7. Strain Should be checked serologically
8. Carrier Should pass through 150-212 micron, IS (72-100 mesh).
9. Others Nodulation test positive, results in 50% or more dry matter yield than control
________________________________________________________________________
Application of inoculants
The major goal of legume inoculation is to introduce efficient and competitive strains in large numbers that can survive and establish in the legume rhizosphere and colonize the roots promptly. Application of inoculant to the seed surface prior to sowing is the traditional, most commonly used and most user-friendly means of inoculation. There are numerous adhesives like gur, sugar, gum arabic and methyl cellulose suitable for attaching inoculant to the seeds.
The method of seed inoculation includes preparation of 10% sugar or pharmaceutical grade gum arabic or 1% methyl cellulose solution. This solution is sprinkled on the seeds and the seeds are thoroughly mixed so as to have a uniform coating. A count of 1000 viable cells per seed is to be attained at the time of treating the seed and quantity of culture used is accordingly adjusted. The seeds are spread uniformly for drying on a gunny bag or cement floor in shade avoiding direct sunlight.
Response of legumes to Rhizobium inoculation
Rhizobium inoculation improves the productivity of leguminous crop plants. The efficacy of Rhizobium inoculation has been established in our country beyond any doubt by the results of coordinated trials conducted by the Indian Council of Agricultural Research. The yield response varies with the inoculant strain, location and crop variety. Average increase in yield of some of the pulse crops due to Rhizobium inoculation is presented in table 8.5.
Percent increase in yield of some leguminous crops due to Rhizobium inoculation
| Crop | % increase |
Arhar | 32 |
Mungbean | 33 |
Chickpea | 41 |
Groundnut | 49 |
Lentil | 50 |
Soybean | 61 |
Azotobacter: Azotobacter is a free living, heterotrophic nitrogen fixing bacteria that occurs in the rhizosphere of variety of plants. The genus Azotobacter has six species viz., A. chroococcum, A. vinelandii, A. beijerinckii, A. nigricans, A. armeniacus and A. paspali. Except the last species, which is a rhizoplane bacterium, the other members are largely soil borne and rhizospheric. The potential of A. chroococcum and A. paspali as a biofertilizer for various non-legume crops is well documented.

Azotobacter is an aerobic, Gram negative, rod shaped bacteria occurs singly, in chains, or in clumps. It does not form endospores but do form thick-walled cysts. These cysts are resistant to desiccation and to some deleterious chemical and physic agents. They, however, cannot withstand extreme temperatures. While in the cyst stage of their life cycle, they do not fix nitrogen and are optically refractile. It may be motile by peritrichous flagella or non-mobile. It can produce a water soluble pigment, either yellow-green, fluorescent or red-violet/ brownish-black. It grows well at an optimum temperature range between 20 and 30°C and at pH 7.0 – 7.5. They are able to grow on various carbohydrates, alcohols, and organic acids.
Azotobacter was first discovered using a culture that was devoid of a combined nitrogen source. Azotobacter is found on neutral to alkaline soils, in aquatic environments, in the plant rhizosphere and phyllosphere. A.chroococcum is the most common species of Azotobacter present in the soil.
Azotobacter sp. are known to influence plant growth through their ability to fix molecular nitrogen; production of growth promoting substances like IAA, gibberellin or gibberellin-like compounds and vitamins, excretion of ammonia in the rhizosphere in the presence of root exudates; production of anti-fungal metabolites and phosphate solubilization.
The procedure followed for mass multiplication of Azotobacter, preparation of carrier based inoculant and seed inoculation with carrier based Azotobacter is similar to that of rhizobial inoculation. Jensen’s N-free medium is routinely used for the mass multiplication of Azotobacter. Seed inoculation of A. chroococcum increases the yield of field crops by about 10 % and of cereals by about 15-20%. The response to inoculation was increased by manuring or by fertilizer application. Coinoculation of Azotobacter with other bioinoculants like Rhizobium; Azospirillum, P-solubilizers; vesicular-arbuscular mycorrhiza have been reported to enhance the growth and yield of legumes, cereals and vegetable crops.
Beneficial effects of Azotobacter chroococcum inoculation has been reported by various workers on various cereal, vegetables, oil seed, legume and cash crops (Table ). Inoculation experiments with Azotobacter gave better yield performance only at lower levels of nitrogen (0 to 30 kg N ha-1). These diazotrophic bacteria require large amounts of available carbon for their survival in soil. Addition of farmyard manure (FYM), compost and other organic amendments to agricultural soils improves the efficiency of Azotobacter and hence the plant grown and yield.
Effect of Azotobacter on crop yield
| Crop | Increase in yield over yields obtained with chemical fertilizers (%) | Crop | Increase in yield over yields obtained with chemical fertilizers (%) |
Food grains | Other | ||
Wheat | 8-15 | Potato | 13 |
Rice | 5 | Carrot | 16 |
Maize | 15-20 | Cauliflower | 40 |
Sorghum | 15-20 | Tomato | 2-24 |
Cotton | 7-27 | ||
Sugarcane | 9-24 |
Source: Das, H.K 1991. Biological nitrogen fixation in the context of Indian agriculture. Curr Sci, May 25, 551-555.
Azospirillum
Beijerinck in 1925 reported a nitrogen-fixing bacterium under the name Spirillum lipoferum. The ability to fix nitrogen by certain spirilla was first recorded by him, who noticed their presence in enrichment cultures of Azotobacter chroococcum. A new orientation to the study of this bacterium has come with the observations of Dobereiner and Day (1976) that Azospirillum could be isolated from the roots of tropical grass Digitaria decumbens using a semi-solid N2-free sodium malate enrichment medium. Surface sterilization of roots by 70% alcohol and creation of micro-aerophilic (low oxygen requirements) conditions in the medium are the two essential steps for the isolation of the organism. Azospirillum is recognized as a very ubiquitous soil organism capable of colonizing effectively not only the roots of a wide variety of plants but also their above ground portions forming apparently an associative symbiosis.
The bacterium is Gram-negatiave, motile, generally vibroid in shape and contains poly-b-hydroxy-butyrate granules. It is very motile and possess a long, polar flagellum for swimming and occasionally, peritrichous flagella for swarming on surfaces. The cells change shape and size with culture age, and produce cysts. They can grow under anaerobic (NO3- as acceptor of electrons, denitrification), microaerophilic (N2 or NH3 as nitrogen sources) and fully aerobic conditions with combined nitrogen only (NH3, NO3-, amino acids). Azospirillum species grow well on organic acids such as malate, succinate, lactate and pyruvate. On Rojo-Congo red medium, Azospirillum forms distinct scarlet red, dry and wrinkled colonies .
Taxonomy
Azospirillum belongs to group 1 of the alpha subclass of the Proteobacteria . At present there are five known species of Azospirillum- A. brasilense, A. lipoferum, A. amazonense, A. halopraeferens and A. irakense. The distinguishing morphological and biochemical characteristics of the five species is given in table .
Different morphological and biochemical characteristics of five known species of Azospirillum
________________________________________________________________________
Characteristics A.lipoferumA.brasilense A.amazonenseA.irakense A.halopraeferens
________________________________________________________________________
Colony type
On CR medium Scarlet Scarlet Pink Scarlet Pink
On PDA medium Pink Pink White White No
growth
Raised Raised Raised Flat Raised
Biotin requirement. + – – – +
Utilization of C
Malate + + + + +
D-Glucose + – – – +
Glycerol + + – – +
Sucrose – – + + –
________________________________________________________________________
Inoculant production
For mass multiplication of Azospirillum, the organism is allowed to grow in flasks containing NH4Cl and malate medium and incubated at 35o – 37o C for 3 days. When there is good growth, the broth culture is mixed with the carrier, and the carrier-based culture is packed in polyethylene pouches. The technique used for preparation of carrier based inoculant and for inoculating the seed or seedlings with Azospirlllum culture is same as that described earlier in case of Rhizobium.
Crop response to Azospirillum inoculation
Azospirillum is extensively used as an inoculant for crop plants belonging to the family gramineae like wheat, sorghum, pearlmillet, fingermillet, barley and maize. Of all the crops tested, sorghum (Sorghum bicolor), pearlmillet (Pennisetum americanum) and fingermillet (Eleusine coracana) appeared to be consistently responsive to Azosprillum at more than one location in India. Azospirillum species promote the yield of agriculturally important crops in many different soils and climatic regions. By the use of this organism as a seed inoculant, savings of 20-30 kg N/ha equivalents can be achieved in these crops. However, the principal effects of azospirilla go far beyond furnishing nitrogen to host plants.
Once inoculated onto plant roots, Azospirillum cells induce remarkable changes in the morphology and behaviour of the entire root system. For instance, hairs close to the root tip take on a more distinctive appearance, and the overall density and the length of the root system increases (Fig ). Root hairs consist of expanded root epidermal cells, which play a role in water and nutrient exchanges and also help to anchor root to its surroundings. Inoculating azospirilla onto plant roots also increases the diameter and length of both lateral and adventitious roots and typically leads to additional branching of the lateral roots. These developments in the root system are important because they increase absorptive area and volume of the soil substrate available to the plant.

Strains of Azospirillum are known to produce siderophores. They are low molecular weight iron binding compounds synthesized in large amounts and excreted into culture medium by microorganisms under iron-deficient conditions. Siderophores form complexes with the metal ions in the culture medium followed by translocation of the complex through bacterial envelope. The ability of Azospirillum to synthesize siderophores may contribute to improve the iron nutrition of plants and offer protection from minor pathogens.
Biosynthesis of growth promoting substances like phytohormones, vitamins, antibacterial and anti fungal substances by Azospirillum is well documented. The most extensively reported growth promoters are IAA, gibberellins, cytokinin like-substances and vitamins.
The ability of azospirilla to form antibiotic substances varies from strain to strain. Fungistatic activity of azospirilla against a wide range of phyto-pathogenic fungi has been reported e.g. certain azospirilla offer protection to cotton plants against Thielaviopsis basicola and Fusarium oxysporum.
These enhancing features of Azospirillum inoculation are also evident in field experiments, with the bacteria not only increasing root numbers but also improving yields of crops such as wheat, sorghum, pearlmillet and maize. In field experiments in Israel, Azospirillum inoculated sorghum plants made better use of moisture stored in soils from winter precipitation than did uninoculated plants. In both green house and field experiments, inoculated plants are efficient at absorbing nitrogen, phosphorus, potassium and other microelements from soil than uninoculated plants.
In recent years, interest has shifted from plant-microbe interaction to plant-microbe-microbe interactions. Several reports have brought to light instances where beneficial effects of Azospirillum on plants are enhanced when coinoculated with other microorganisms like Rhizobium and Azotobacter. Synergistic effects of Azospirillum with Rhizobium on various legumes have been reported. Stimulation of nodulation may be due to an increase in production of lateral roots and in root hair branching. This, in turn, has been thought to be due to production of phytohormones by Azospirillum. The positive effect of inoculating non-legumes with Azospirillum brasilense and Azotobacter chroococcum, at low application rates of mineral N, on associative N2 fixation and on crop yield has been reported.
Acetobacter diazotrophicus
Acetobacter diazotrophicus, is a gram-negative, microaerobic, nitrogen fixing microorganism and was isolated from washed roots and stems of sugarcane, using semi-solid N-free sugar medium acidified with acetic acid to pH 4.5. Cells of Acetobacter diazotrophicus are straight rods with rounded ends, about 0.7 to 0.9 by 2 um, motile by lateral or peritrichous flagella. Optimum growth temperature is around 30C. Although sucrose is the best C-source for Acetobacter diazotrophicus but sugars like glucose, fructose, galactose, mannitol are also utilized. It grows well in the pH range of 3.8 to 5.8 with good nitrogenase activity. Growth and nitrogen fixation occur at sugar concentration ranging from 10 to 30 %.
Acetobacter diazotrophicus, an endophytic diazotroph, has been found mainly associated with sugar-rich plants such as sugarcane, sweet potato, Cameroon grass, sweet sorghum and coffee. It colonizes roots, stems and leaves of host plants. Reports from Brazil indicates that A. diazotrophicus contributes >50% of biologically fixed nitrogen in sugarcane. Nitrogen fixed by A. diazotrophicus is excreted as ammonia into the medium. Strains of Acetobacter have been shown to produce considerable amount of IAA. Synergistic effects on plant growth and yield following inoculation with Acetobacter diazotrophicus and AM fungi have been reported for sugarcane, sweet potato and sweet sorghum.
Blue green algae and Azolla
Blue-green algae (cyanobacteria) are ubiquitous in distribution. They are either single celled or consist of branched or unbranched filaments (Fig. a,b,c). It is a group of free living organisms that has been demonstrated to be an ideal candidate as the biological nitrogen source in rice ecosystems. Some of them possess a peculiar structure known as ‘heterocyst’ and all heterocystous forms can fix nitrogen from air. Recently, some blue-green algae without heterocysts have also been found to fix nitrogen under special conditions like low oxygen tension. The algae that are generally used for field application are species of Aulosira, Tolypothrix, Scytonema, Nostoc, Anabaena and Plectonema as a mixture.

Cyanobacteria have ability to carry out both photosynthesis and nitrogen fixation. Besides contributing to the nitrogen economy of the soils these algae have other beneficial effects. Their exceptionally good water holding capacity, their ability to concentrate nutrients such as nitrogen, phosphorus, fixed carbon and trace elements, their soil binding capacity and their ability to scavenge sodium from salt affected soils are additional ecological advantages. The presence of BGA in the immediate vicinity of rice seeds can decrease sulphide and iron injury to the plants. Cyanobacteria also produce number of plant growth substances like amino acids, small proteins and peptides, sugars, complex polysaccharides, vitamins and growth hormones. Standing crops of nitrogen fixing BGA range from 5-20 tons per hectare fresh weight and contribute approximately 30kg nitrogen per season per hectare of rice field. A bulk of the organic matter produced by algal growth remains in the soil and becomes available to the next crop as organic enrichment
Production of algae for field application
Based on the natural ecology of these algae, a simple rural-oriented open-air method of producing them in bulk has been developed. The basic principle is to grow them in natural sunlight under conditions stimulating these in the rice field. You can use a starter culture, consisting of soil-based mixture of efficient strains of BGA, supplied by various agricultural universities for mass multiplication.
Shallow stays (15cm x 7.5 cm x 22.5 cm) of galvanized iron sheet, or brick and mortar, or pits lined with polythene sheets are prepared. The size can be increased if more material is to be produced. About 10kg soil is placed and mixed with 200 g super phosphate. The trays are then filled with water (5-15 cm) depending upon the local conditions and rate of evaporation; the pH of the soil should be around neutral. After soil settles down, saw-dust and the starter culture are sprinkled on the surface of the standing water. The whole assembly is exposed to sunlight. In hot summer months, the growth of the algae will be rapid and in about a week a thick algal mat will be formed on the surface of the soil and sometimes even floats up. If the daily rate of evaporation is high, more water is added intermittently. When the algal growth becomes sufficiently thick, addition of water should be discontinued and the water is allowed to dry up in the sun. The dried algal flakes are collected from the surface and stored in bags for future use in the fields. The trays are again filled with water and a small amount of the dry algal flakes is added, as further inoculum. The process is continued as above. Once the soil in the tray is exhausted (usually 3-4 harvests), fresh soil is put and mixed with super phosphate and the process is repeated as before. To prevent the breeding of insects, application of Malathion (0.00075 ppm) or Carbofuran (3% granules) is recommended.
Algae are applied at the rate of 10kg/ha over the standing water in the field one week after transplantation. The field is kept waterlogged at least for a couple of days immediately after algal application.
Azolla-Anabaena symbiosis
Azolla is a small aquatic fern and is omnipresent in nature. Each leaf consists of two lobes, a thick aerial dorsal lobe and a thin ventral lobe occasionally of a slightly larger size. The dorsal lobe is green and has a blue green algal symbiont (Anabaena azollae) within a central cavity. The heterocyst of the symbiont Anabaena is the site of nitrogen fixation. Azolla provides nutrients and a protective leaf cavity for Anabaena, which in turn provided nitrogen for the fern.
Azolla is found on still water in ponds, lakes, swamps, ditches and paddy fields of temperate and tropical regions. Because of its rapid growth, high N content and ability to grow in still water, it has been exploited as a fertilizer for rice. This is used in Vietnam and China for centuries, however, its use as a biofertilizer in India is relatively a recent development. There are 7 living species of Azolla – A. pinnata, A. caroliniana, A. rubra, A filiculoides, A. nilotica, A. mexicana and A. microphylla. A. pinnata is native to India but now many of these species have been introduced.
The high N2 fixing ability, rapid growth, high biomass accumulation and N-content determines the potential of Azolla as a biofertilizer for rice. Biological nitrogen fixation through Azolla – Anabaena complex is considered a potential biological system for increasing rice yield at comparatively low cost. The ability of Azolla to fix N2 is about 1.1 kg N/ha/day. The doubling time varies between 2 and 10 days for most species and maximum biomass ranged between 0.8 to 5.2 t dry matter/ha with an average of 2.1 t/ha.
Large scale production of Azolla
The potential Azolla species are maintained in concrete tanks keeping soil under flooded conditions. Partial shade helps during summer months. From these Azolla is harvested and used as inoculum in bigger size plots or in small ponds generally found in villages of rice growing areas.
Its large-scale production is carried out in a nicely prepared field divided into small sub-plots with good irrigation facility (4-50 sq.m. plot with 5-10 cm water depth). Azolla is inoculated at the rate of 0.5 to 1.0 t/ha. Inoculation with higher doses ensures rapid multiplication. Super phosphate at the rate of 4-8 kg/ha stimulates fern growth. Insecticide like Furadan is also applied (2.5-3.0 kg/ha). Under optimum conditions, Azolla forms a thick mat on water surface in 15-20 days. About two-third of it is harvested and the remaining is left for further multiplication. It again multiplies and forms a thick mat in 2-3 weeks. About 100kg fresh Azolla inoculum can be obtained every week from a nursery of 100m2. Super phosphate at the rate of 60 kg/ha can be split into 2-3 doses or added at week interval to have better results. If Azolla multiplication is good even without addition of P, then there is no need to add it.
Phosphate solubilizing and mobilizing microbes
Phosphorus is a major nutrient required for the growth of plant. There are large reserves of phosphorus in soils but very little amount is available to the plant. There are microorganisms in soil that can solubilize the unavailable phosphorus and make it available to plant. They are called Phosphate solubilizing microorgamisms (PSM). A group of fungi associates with the roots of higher plants and mobilize the phosphorus from soil to the plant system.
Phosphate solubilizing microorganisms
The majority of agricultural soils contain large reserves of phosphorus of which a considerable part has accumulated as consequence of regular applications of P-fertilizer. The phenomenon of fixation and precipitation of P in soil, which is highly dependent on pH, causes a low efficiency of soluble P fertilizers. In acidic soils P is precipitated as Al and Fe phosphates whereas in calcareous soils high concentration of Ca results in P precipitation. The soil is a habitat for diverse group of organisms that employ variety of solubilization reactions to release soluble phosphorus from insoluble phosphates. The potential of these phosphate solubilizing microorganisms has been realised and are utilised as bioinoculants for crop grown in soils poor in available P and amended with rock phosphate or tricalcium phosphate.
Phosphorus solubilizing microorganisms include various bacterial, fungal and actinomycetes forms which help to convert insoluble inorganic phosphate into simple and soluble forms. Members of Pseudomonas, Micrococcus, Bacillus, Flavobacterium, Penicillum, Fusarium, Sclerotium and Aspergillus are some of the phosphate-solubilizing micro-organisms. They normally grow in a medium containing insoluble tri-calcium phosphate [Ca3(PO4)2], apatite, rock phosphate, FePO4 and AIPO4 as sole source of phosphate. The initial isolation of phosphate solubilizers is made by using Pikovaskaya medium suspended with insoluble-phosphates such as tri-calcium phosphate. The production of clearing zones around the colonies of the organism is an indication of the presence of phosphate-solubilizing organisms (Fig. ). Such cultures are isolated, identified and the extent of solubilization determined quantitatively. Several rock phosphate dissolving bacteria, fungi, yeast and actinomycetes were isolated from soil samples collected from rock phosphate deposits and rhizosphere soils of different leguminous crops. The most efficient bacterial isolates were identified as Pseudomonas striata, Pseudomonas rathonis and Bacillus polymyxa and fungal isolates as Aspergillus awamori, Penicillium digitatum, Aspergillus niger and a yeast-Schwanniomyces occidentalis. These efficient micro-organisms have shown consistently their capability to solubilize chemically-fixed soil phosphorus and rock phosphate from different sources – Mussorie, Udaipur, Matoon, Singhbhum, Morocco, Gafsa and Jordan. In addition, these micro-organisms were found to mineralize organic phosphorus to soluble form due to enzymatic activity.

The efficient cultures have shown capacity to solubilize insoluble inorganic phosphate such as rock phosphate, tri-calcium phosphate, iron and aluminium phosphates by production of organic acids. They can also mineralize organic phosphatic compounds present in organic manure and soils. Inoculation of PSM to seeds or seedlings increases the grain yield of crops. They are known to add 30-35 kg P2O5 ha-1.
The inorganic phosphate solubilization by microbes can be attributed to acidification, chelation, exchange reaction in growth medium as well as to the proton transfer during ammonium assimilation.
Phosphate mobilizing microbes: Mycorrhizae
The term mycorrhizae was coined for symbiotic associations formed by fungi with roots (Gr. myces = fungus, rhizo = roots). Mycorrhizae are wide spread under natural conditions and occur nearly in all soils from mine spoils to agricultural soils as well as soil under horticultural or fruit crops. More than 95% of plant taxa form mycorrhizal associations. The association is generally mutualistic in that the fungi obtain a carbon source from host, whilst the latter benefits from enhanced nutrient uptake through transfer from soil via the fungi. They are formed by most vascular plants except for a few monocotyledons like cyperacea or juncacae and dicotyledons like chenopodiacea or brassicaceae. Mycorrhizea are usually divided into three morphologically distinct groups depending on whether or not there is fungal penetration of root cells : endomycorrhiza, ectomycorrhiza and ectoendomycorrhiza. Of the three groups, endomycorrhizae are important as biofertilizer.

Endomycorrhizae are formed by nearly 90% of the land plants. In this association the fungi form external hyphal networks in the soil and grow extensively within the cells of the root cortex. This network of fungal hyphae within the root cortex is known as hartig net. Fungi belonging to basidiomycetes, ascomycetes or zygomycetes are involved depending on the type of endomycorrhizal association. Specific types of endomycorrhizae are formed by members of the Ericaceae (Ericoid mycorrhizae) and orchidaceae (orchidaceous mycorrhizae), but the type of mycorrhizae which is widespread is the arbuscular mycorrhizae (earlier referred as vesicular-arbuscular mycorrhizae). It is formed by 120 species of zygomycetes, all belonging to the order Glomales (Glomus, Acaulospora, Gigaspora, Sclerocystis, Entrophospora and Scutellospora). None of these fungi has yet been successfully cultured axenically.

The effect of mycorrhizae in increasing plant growth has been well documented by different workers for many plants. The beneficial effect of mycorrhizae on plant growth has mostly been attributed to an increase in the uptake of nutrients, especially phosphorus. Mycorrhizal fungi improve the soil phosphorus availability by solubilizing inorganic forms of phosphorus or by mineralization of organic phosphorus. External hyphae of mycorrhiza also has the capacity to take up and deliver various other nutrients to plants like NH4+, NO3- ,K, Ca, SO42- , Cu, Zn and Fe. In experimental chambers, the external hyphae of AM can deliver upto 80% of plant P, 25 % of plant N, 10% of plant K, 25% of plant Zn and 60% of plant Cu. Mycorrhiza also produce ectoenzymes which provide host plant with the potential to access organic N and P forms that are normally unavailable to AM fungi or to non mycorrhizal roots.
Plant Growth Promoting Rhizobacteria (PGPR)
The environment, or the volume of soil that is influenced biologically and biochemically by living root, is known as rhizosphere. Root exudates and secretions create a rhizosphere effect that manifests itself in the intense microbial activity that is associated within the immediate vicinity of the root. Root associated bacteria, also called rhizobacteria , can be beneficial, neutral or deleterious to the growth of the plant. Plant growth promoting rhizobacteria (PGPR) are one class of beneficial bacteria inhabiting the soil ecosystem (Kloepper et al 1989). The effects of PGPR on plant growth can be mediated by direct or indirect mechanisms (Glick 1995). The direct effects have been most commonly attributed to the production of plant hormones such as auxins, gibberellins and cytokinins, or by supplying biologically fixed nitrogen or solubilizing insoluble P. These PGPR also affect growth by indirect mechanisms such as supperssion of bacterial, fungal and nematode pathogens by the production of siderophores, HCN, ammonia, antibiotics, volatile metabolites etc., by induced systemic resistance and/or by competing with the pathogen for nutrients or for colonization space.
The nitrogen fixing and phosphate solubilising bacteria have been discussed separately. Other PGPR include bacteria belonging to the genera Arthrobacter, Bacillus, Burkholderia, Enterobacter Klebsiella, Pseudomonas, Xanthomonas, Serratia and many more yet to be identified. Effects of PGPR on plant growth have been evaluated by many workers on different crops. Increase in plant height and root and shoot biomass of wheat was reported following inoculation with 12 different isolates of PGPR belonging to Pseudomonas aeruginosa, P. cepacia, P. fluorescens and P. putida. Similarly treatment of wheat seeds with fluorescent pseudomonads (antagonistic to Gaeumannomyces graminis) resulted in yield increases of 27% in field trials. PGPR are potent inoculants but are not commercialised due to lack of consistency under field conditions. Recent work suggests that combination of PGPR strains (two or more) which have diverge mode of plant growth promotion or antagonism against soil-borne pathogens are more effective than single strain inoculum. IAA producing Bacillus isolates promoted root growth and (or) nodulation when coinoculated with Rhizobium etli on Phaseolus vulgaris. Similarly coinoculation of soybean with B. japonicum and Serratia liquefaciens 2-68 or S. proteamaculans 1-102 increased soybean grain and protein yield compared to the non treated controls. Better biocontrol of take all disease of wheat was observed when fluorescent Pseudomonas was applied in combination with Trichoderma koningii. Seed treatment containing combinations of Escherichia coli S17R1 and Burkholderia cepacia BC-B provided significantly greater suppression of cucumber seedling pathogenesis in a field soil naturally infested with Pythium and Fusarium spp. than seeds treated with strain BC-B, S17R1 or Enterobacter cloacae 501 R3. Experiments performed at chernobyl showed that coinoculation of ‘duet’ of nitrogen fixing Klebsiella oxytoca VN13 and Xanthomonas maltophila VN12 could protect maize from radionuclides penetration; as well improve the yield and percentage of protein in seed.
Many bacterial genera have shown their potential for biocontrol both under in vivo and in vitro conditions. Agrobacterium, Arthrobacter, Alcaligenes, Bacillus, Escherichia coli, Enterobacter, Pseudomonas, Burkholderia, Rhizobium and Serratia were found to be potent for suppression of soil-borne fungal pathogens. Many of these biocontrol agents exhibited their effectiveness under field conditions also.
| Download this lecture as PDF here |
The intensive use of pesticides in agriculture is a cause of serious concern. The problem is especially serious because of the development of resistance to pesticides in important pests and the presence of pesticide residue in agricultural and dairy products. In India, the most serious problem of resistance is witnessed in cotton, for which American bollworm is a serious pest. The bollworm has developed resistance to almost all pesticides in a number of regions and is serious problem in many states. Other important pests of cotton, white fly and jassid, have also developed pesticide resistance in some places. Growing pesticide resistance has meant that a large proportion of agricultural production is lost to pests. Pesticide resistance has mainly been caused by excessive and indiscriminate use of pesticides. Pesticides of spurious quality, which are commonly sold in small towns and villages, have also contributed to resistance in many areas.
Excessive use of chemical pesticides in agriculture is a serious cause of concern. It is, therefore, important that alternative, environmental friendly methods of plant protection are adopted such as integrated pest management (IPM) techniques, including the use of biopesticides.
Biopesticides are an important group of pesticides that can reduce pesticide risks. They are derived from animals, plants and microorganisms such as bacteria and viruses. The advantages of biopesticides are:
- They are inherently less harmful than chemical pesticides.
- They, in general, have a narrow target range and a very specific mode of action.
- They are often effective in small quantities. Also, they decompose quickly and do not leave problematic residues.
- They are safer to humans and the environment than conventional pesticides.
Biopesticides is a broad term and includes bioinsecticides, biofungicides, bioherbicides and bionematicides. Microorganisms belonging to different groups like bacteria, fungi ans viruses are used as biopesticides. You will learn about these three groups of organisms in following text.
Bacterial
Bacteria belonging to genus Bacillus are potent against many insect pests. They suppress pests by producing a toxin specific to the pest; causing a disease; preventing establishment of other microorganisms through competition; or other modes of action.
An example of a bacterial pesticide is Bacillus thuringiensis, or “Bt.” Bacillus thuringiensis is a naturally occurring soil bacteria that is toxic to the larvae of several species of insects but not toxic to non-target organisms. It is primarily a pathogen of lepidopterpous pests that are some of the most damaging. These include american bollworm in cotton and stem borers in rice. Bacillus thuringiensis can be applied to plant foliage or incorporated into the genetic material of crops. Bacillus thuringiensis, as discovered, is toxic to the caterpillars (larvae) of moths and butterflies. Several strains of Bt have been developed and now strains are available that control fly larvae. These can be used in controlling mosquitoes and blackflies.
Bacillus thuringiensis (Bt) is a ubiquitous gram-positive, spore forming bacterium which produces parasporal crystals during sporulation (stationary phase of its growth cycle). These crystals are predominantly comprised of d-endotoxins or insecticidal crystal proteins (ICPs), known to possess insecticidal activity when ingested by certain insects. The mode of action of Bt involves the following stages:
- ingestion of sporulated Bt and ICP by an insect larva.
- solubilization of the crystalline ICP in the midgut: When Bt crystals are ingested by insects, the crystal proteins are dissolved from the crystals. The pH in the gut of lepidopteran larvae varies between 9 and 12 and the lepidopteran-specific crystal bodies can only be solubilized above pH9.5. On getting solubilized in the midgut, the crystalline bodies release the protein called d-endotoxins.
- activation of the ICP by midgut proteases: The crystalline protoxins are inactive, until they are hydrolysed by the gut proteases. The proteases cleave amino acids from both C-terminus and N-terminus of the protoxin and thus forms the active toxin.
- binding of the activated ICP to specific receptors in the midgut cell membrane: Brush border membrane vesicles (BBMVs) is the primary binding site for several insect species. The active toxins initially bind reversibly to the specific receptors located on the apical brush border membrane of the columnar cells.
- insertion of the toxin in the cell membrane and formation of pores and channels in the gut cell membrane, followed by destruction of the epithelial cells: After binding to the receptor, the toxin inserts irreversibly into the plasma membrane of the cell. The formation of toxin induced pores in the columnar cell of apical membranes allows rapid fluxes of ions. The disruption of the gut integerity leads to the death of the insect through starvation or septicemia.
- subsequent Bt spore germination and septicemia may enhance mortality.
For biopesticide applications, the Bt protein is usually used in a formulation containing the spores and crystalline inclusions that are released upon lysis of Bt cells during growth. The molecular potency of the toxin is 300 times greater than synthetic pyrethroids, and the toxin breaks down quickly when exposed to ultraviolet light/sunlight.
Besides Bacillus thuringiensis, other bacteria like Bacillus popilliae and B. sphaericus are also important for their biocontrol activity. B. popilliae is a Gram-negative spore-forming rod, 1.3 to 5.2 x 0.5 to 0.8 micrometres. It is a fastidious organism that grows only on rich media containing yeast extract, casein hydrolysate or an equivalent amino acid source, and sugars. Trehalose, the sugar found in insect haemolymph, is a favoured carbon source though glucose also can be used. Some varieties of B. popilliae form a crystalline body inside the cell at the time of sporulation and in this respect resemble B. thuringiensis. But the crystal is not thought to play a significant role in infection and certainly it is not as important as in B. thuringiensis. The variety lentimorbus, for example, does not produce a crystal and yet it causes disease. Another difference between B. popilliae and B. thuringiensis is that B. popilliae cannot be induced to sporulate in laboratory media although it does so readily in the diseased host. Actually there are a number of oligosporogenic mutants – ones that produce a few spores – but spores for microbial control programmes are usually produced in living insect larvae – an expensive and time-consuming process. Its spectrum of control includes larvae of Japanese beetles, chafers, some May and June beetles. Spores of B. popilliae persist for long periods in the soil and are ingested by grubs in the soil, and multiply in the hemocoel. The infected larvae do not molt to the next instar, remain active until just prior to death when they become sluggish and moribund.
Bacillus sphaericus is also used to control specific kinds of mosquitoes (especially Culex), including some that transmit diseases such as encephalitis. It is active against the larvae of Culex, Psorophora and Anopheles species; less effective against Aedes species. It is a naturally occurring bacteria – isolated, cultured, and labeled for mosquito control. Bacillus sphaericus acts as an endotoxin to mosquito larvae. It is consumed by the larvae as live bacterium. The bacterium is able to penetrate through the intestines of the mosquito larvae into the hemocoel. Once in the hemocoel, B. sphaericus reproduces and releases lethal doses of toxin killing the mosquito larvae.
Fungal
Beauveria is a naturally occurring fungus in soils throughout the world, and has been researched for control of soil borne insects. Many soil insects, however, may have a natural tolerance to this pathogen, which is not exhibited in many foliar pests. Therefore, commercial development of this fungus for biological control has primarily been targeted against foliar feeding pests.
Beauveria bassiana causes a disease known as the white muscadine disease in insects. Beauveria belongs to fungal subdivision: Deuteromycotina and order: Hyphomycetes. It has a simple life cycle with no known sexual stage; the asexual spores are called conidia. Many strains of Beauveria bassiana are used as biopesticides. It is active against adults and larvae of many kinds of insects; eggs of lepidopteran pests such as moths. The spectrum also includes mole cricket, chiggers, white grubs, fire ants, ants, flea beetle, boll weevil, whiteflies, plant bug, grasshoppers, thrips, aphids, mites and many others.
When spores of this fungus come in contact with the cuticle (skin) of susceptible insects, they germinate and grow directly through the cuticle to the inner body of their host. The fungus proliferates throughout the insect’s body, producing toxins and draining the insect of nutrients, eventually killing it. Therefore, unlike bacterial and viral pathogens of insects, Beauveria and other fungal pathogens infect the insect with contact and do not need to be consumed by their host to cause infection. Once the fungus has killed it’s host, it grows back out through the softer portions of the cuticle, covering the insect with a layer of white mold (hence the name white muscadine disease). This downy mold produces millions of new infective spores that are released to the environment.
Viral (Insect Viruses)
Baculoviruses are pathogens that attack insects and other arthropods. Like some human viruses, they are usually extremely small (less than a thousandth of a millimeter across), and are composed primarily of double-stranded DNA that codes for genes needed for virus establishment and reproduction. Because this genetic material is easily destroyed by exposure to sunlight or by conditions in the host’s gut, an infective baculovirus particle (virion) is protected by protein coat called a polyhedron (plural polyhedra). Most insect baculoviruses must be eaten by the host to produce an infection, that is, typically fatal to the insect.
The majority of baculoviruses used as biological control agents are in the genus Nucleopolyhedrovirus. These viruses are excellent candidates for species-specific, narrow spectrum insecticidal applications (Table 8.8). They have been shown to have no negative impacts on plants, mammals, birds, fish, or even on non-target insects.
On the other hand, the high specificity of baculoviruses is also cited as a weakness for agricultural uses, since growers may want one product to use against a variety of pests. Currently, researchers are attempting to use genetic engineering techniques to expand virus host ranges to the desired pest species. Releases of such genetically-engineered baculoviruses have been made by researchers in the U.K. and the United States and show promise, although the cost of commercial production of these agents must be reduced if they are to be competitive.
Viruses are unable to reproduce without a host – they are obligate parasites. Baculoviruses are no exception. The cells of the host’s body are taken over by the genetic message carried within each virion, and forced to produce more virus particles until the cell, and ultimately the insect, dies. Most baculoviruses cause the host insect to die in a way that will maximize the chance that other insects will come in contact with the virus and become infected in turn. Infection by baculovirus begins when an insect eats virus particles on a plant – perhaps from a sprayed treatment. The infected insect dies and “melts” or falls apart on foliage, releasing more virus. This additional infective material can infect more insects, continuing the cycle.
It is widely acknowledged that baculoviruses can be as effective as chemical pesticides in controlling specific insect pests. However, the expense of treating a hectare of land with a baculovirus product invariably costs more than an equally efficacious chemical treament. This difference in price is due primarily to the labor intensive nature of baculovirus production. Some viruses can be produced in vitro (within cell cultures in the laboratory, not requiring whole, living insects). These are less expensive than those that can only be produced in vivo, that is, inside of living insects. The cost of rearing live hosts adds greatly to the final cost of the product. It is to be hoped that insect cell culture systems currently being developed for other uses may ultimately make viral pesticides more cost-effective.
Insects killed by baculoviruses have a characteristic shiny-oily appearance, and are often seen hanging limply from vegetation. They are extremely fragile to the touch, rupturing to release fluid filled with infective virus particles. This tendency to remain attached to foliage and then rupture is an important aspect of the virus life-cycle. As discussed above, infection of other insects will only occur if they eat foliage that has been contaminated by virus-killed larvae.
Viruses used against different insect-pests of plants

Biocontrol
Biocontrol or Biological control can be defined as the use of natural enemies to control pests. Natural enemies of pests are categorized as parasites, predators and pathogens. It is a broad term that also includes use of biopesticides.
About 30% of the yield in agriculture is lost because of the combined effects of biotic and abiotic stresses, with pathogenic fungi alone responsible for a reduction of about 12%. Control of fungal pathogens is based on the use of agronomic practices and pesticides, but widespread application of chemicals inundates the agroecosystems with toxic compounds that affect the balance of the natural food chain. In addition, resistant and more virulent pathogen populations are selected causing escalation in the amount of pesticides used. A variety of new technologies are being developed to integrate or substitute the application of chemicals in an attempt to reduce both the ecological and financial cost of disease control. Antagonistic microorganisms are being studied in depth and considered as an attractive option for the development of microbial-based biofungicides. Successful and consistent results have been achieved with some biocontrol agents such as Agrobacterium and Bacillus, whose mechanisms of biocontrol are largely understood. However, limitations in the practical use of bacterial agents often arose from the production of toxic substances and formulations with a short shelf-life. The application of fungal biocontrol agents has also been delayed because of difficulties in obtaining consistent results in biocontrol and the relatively poor understanding of the plant-microbe and microbe-microbe interactions involved in the antagonistic processes.
While diverse microbes may contribute to the biological control of plant pathogens, most research and development efforts have focused on isolates of three genera, Bacillus, Trichoderma, and Pseudomonas.
The most studied fungal biocontrol agents are Trichoderma spp. and some isolates, effective as biofungicides in certain culture conditions, have been recently introduced in commercial agriculture. Concurrently, fundamental discoveries concerning the mechanism of action of these fungi have been made. Studies on the mechanism of biocontrol had indicated that Trichoderma and other mycoparasites have developed a vast array of molecular tools to support their parasitic behaviour. It is believed that Trichoderma produces different types of lytic enzymes that act on the cell wall of fungi and kill them. Genes encoding for cell wall degrading enzymes (CWDES) such as chitinolytic, glucanolytic and proteolytic enzymes have been isolated and used to improve biocontrol capabilities of Trichoderma strains.
Two species of Trichoderma, T. harzianum and T. viride are commonly used as biocontrol agent. Their spectrum of control includes fungal pathogens like Armillaria, Pythium, Rhizoctonia, Verticillium, Sclerotium and Botrytis.
| Download this lecture as PDF here |
BIOGAS (Methane)
Biogas is mixture of methane (50-60 per cent), CO2 (30 – 40 per cent), hydrogen (5-10 per cent), H2S and nitrogen (traces), produced from anaerobic digestion of animal, plant wastes or any cellulose containing waste material. The digester used for biogas production is called a Biogas plant. A typical biogas plant using cowdung as a raw material consists of: (a) digester and (b) gas holder. The digesters are either of (a) batch type which are filled once, sealed and emptied when the raw materials stop producing gas or (b) continuous type which are fed with a definite quantity of waste at regular intervals so that gas production is continuous and regular. The nature of fermentation in the digester is anaerobic
Methane production
Involves three process viz., hydrolysis, acidification and methonization
- Four different groups of organisms are involved
- Hydrolytic bacteria -catabolises carbohydrates,proteins, lipids and other components of biomass into fatty acids,H2 and CO2
- H2 producing acetogenic bacxteria :Catabolises fatty acids, and the neutral ebd products of I group into acetate,CO2 and H2
- Homoacetogenic bacteria : Synthesize acetate using H2,CO2 and formate
- Methanogenic bacteria : Utilizes acetate,CO2,H2 to produce methane
The first group of bacteria includes facultative as well as strict anaerobes like Cellulomonas, Bacillus, Eubacterium etc.

Methanogenic phase
- It is a strict anaerobic phase and during this phase organic Carbon is converted to microbial mass, CO2 and methane.
- These bacteria are sensitive to pH and optimum is 6.8- 7.2. Drop in pH leads to inhibition of methanogenesis
- Methanobacterium, Methanomicrobium, Methanococcus, Methanosarcina


Alcohol production
Production of ethyl alcohol from sugary materials is one of the oldest known microbiological processes. Alcohol is an important solvent and raw material used in a variety of chemical industries. Although today industrial alcohol is also produced synthetically from ethylene, production of alcohol by fermentation of cheap sugary materials such as molasses by yeast is still an important industry.
For ethyl alcohol production, selected strains of Saccharomyces cerevisae are employed since all the strains are not equally efficient. The alcohol tolerance and sugar tolerance are important criteria used in the selection of yeast strains. Strains tolerant to high sugar and alcohol concentration are desired. The raw material generally used is either crude cane molasses or best molasses which contain about 50 per cent fermentable sugars. The production process involves the dilution of molasses to a suitable sugar concentration (15- 16 per cent sugars), addition of small quantity of nitrogen source (urea, ammonium sulphate or ammonium phosphate), adjustment of pH to about 5.0 and the addition of an actively growing yeast culture. The fermentation is carried out in big deep tanks of steel or stainless steel. The fermentation is allowed to continue for about 24 – 36 h at 250C to 300C after which the cells are allowed to settle. The fermented mash is then distilled and passed through rectifying columns to recover ethyl alcohol. A large amount of carbon-di oxide is also produced during the fermentation which is purified and compressed. The yield of ethyl alcohol is about 50 per cent of the fermentable sugar concentration. Further purification of ethyl alcohol is done by fractional distillation. In some distilleries, the yeast is recovered and used as animal feed while in most, it is discarded into the effluents, a procedure that is very undesirable.
In recent years because of the possibility of using ethyl alcohol as a fuel supplement and a chemical feed stock, there is increased interest in increasing production but at a cheaper and economical rate. For this, a variety of improvements in the traditional batch fermentation have been described in literature. Among these, the one that has attracted attention is the cell recycle technique which does not involve much additional expenditure. Basically, the technique involves the reuse of cell mass that is produced during the fermentation. It has been found that by doing so, about 5-10 per cent of the substrate which would have been otherwise used for cell growth is saved in addition to a great saving in the cost of inoculum and time. By using recycling technology, fermentation time has been drastically reduced from 24- 36 hours in a batch fermentation to as low as 5-6 hours.
Distilled Alcoholic Beverages
Whisky, rum, gin, brandy and vodka, are all products of alcoholic fermentation of different raw materials such as grains, molasses, potatoes and fruits are distilled products containing higher content of alcohol than either wine or beer. In principle, production of these alcoholic beverages is similar to the production of industrial alcohol. The quality of the alcohol produced and used for the production of these beverages depends on the type of secondary fractional alcohol distillation. It is essential to derive a pure spirit for the production of these beverages.
Whisky is traditionally produced from the fermentation of grain mash, rum from molasses, brandy from grape wines and vodka from potatoes. The flavour and aroma are introduced into the purified alcohol during the blending process. Sometimes, these alcoholic beverages are allowed to age for long periods in wooden casks before they are consumed.
| Download this lecture as PDF here |
Introduction
Although plastics as we know them today are a relatively recent invention, they have become an important part of modern life.
The age of plastics
Today, 200 billion pounds (100 million tons) of plastics are produced worldwide every year. Plastics are used for packaging, building materials, and virtually every type of consumer product. Past ages of human society have been called the Stone, Bronze, Copper, Iron, and Steel Ages, based on the material that was relied upon the most during that time. Today, the total volume of plastics produced worldwide has surpassed that of steel and continues to increase. Without a doubt, we have entered the Age of Plastics.
Some common plastic items include: sunglasses, tooth brushes, super glue, paint brushes, tennis shoes, Frisbees, 2-liter bottles, Honda CRX’s, Astroturf, photographs, street signs, pens, automobile paint, video tapes, rubber bands, balloons, bicycle tires, umbrellas, guitar strings, carpeting, shower doors, hearing aids, Scotch Tape, fishing lines, trash bags, and toilet seats. Plastic can be found in everything from clothing to machinary.
It is important to understand the nature of plastics, and the consequences of their production and use. Virtually all plastics are made from nonrenewable resources, such as oil, coal or natural gas, which will eventually become exhausted. Plastics waste is increasing, adding to the already burdensome problems of waste management. And the use of plastics continues to grow, raising the important question, how can we balance convenient living with concern for ecology? To understand this concern, it is helpful to understand what plastics are.
Why green plastics?
Green plastics are the focus of an emerging industry focused on making convenient living consistent with environmental stability. One reason to make a shift toward the use of green plastics is the availability of raw materials. Green plastics can be made using polymers that come from agricultural and marine feedstocks. These are abundant natural resources that are constantly being replenished. This, in turn could revitalize rural economy, both agricultural and marine, by providing additional demand for currently underutilized land or low-valued biomass commodities.
Another favorable property of green plastics is their biodegradability, making them a natural material for use in such applications as compostable collection bags, such as for food or yard waste.
But bioplastics have to possess adequate physical properties. Their properties have to be managed and controlled with technological means through the development of adequate formulations and plastics processing. The commercial ventures already under way in the United States, Canada, Europe, and Japan indicate that there is confidence technological advances are possible. The key to solving technical problems is often simply knowing what the problems are.
Bioplastics also have to be cost-competitive. Commercially available biopolymers are typically more expensive than synthetic polymers, often significantly so. Currently only starch competes with synthetic polymers in terms of cost.
Interest in the development of bioplastics will grow largely to the extent that there is real interest in and concern over the environment. Societal concern over the environment is already being reflected in governmental restrictive legislation on the use of plastics, particularly aimed at plastic packaging. Legislation has begun at the local, state, federal, and international levels, and legislation will undoubtedly increase in the future. New legislation will likely contain restrictions aimed at materials that are neither recyclable nor biodegradable. Labeling legislation may lead to an “ecolabel,” based on a product’s raw material usage, energy consumption, emissions from manufacture and use, and waste disposal impact. Most of all, what is needed is a paradigm shift.
Making it a reality
Ignoring nature’s way of building strong materials, we have, for many applications, over-engineered our plastics for stability, with little consideration of their recyclability or ultimate fate, and ended up transforming irreplaceable resources into mountains of waste.
There is another way. We can take nature’s building materials and use them for our purposes, without taking them out of nature’s cycles. We can be borrowers, not consumers, so that the process can continue indefinitely. If society is indeed, becoming more and more committed to resource conservation, environmental preservation and sustainable technologies, bioplastics will find their place in this Age of Plastics.
The widespread use of these new plastics will depend on developing technologies that can be successful in the marketplace. That in turn will partly depend on how strongly society is committed to the concepts of resource conservation, environmental preservation, and sustainable technologies. There are growing signs that people indeed want to live in greater harmony with nature and leave future generations a healthy planet. If so, bioplastics will find a place in the current Age of Plastics.
Plastics
Plastics are a class of material that has one or more polymers as its primary ingredient, that is shaped by flow when it is processed (usually using heat), and that is solid in its final form. Plastics can be made up of many different kinds of polymer, and can be processed in many different ways, but as long as they satisfy these three conditions, they are bona fide plastics.
The general “recipe” for any kind of plastic is a combination of three ingredients: a polymer, one or more plasticizers, and one or more additives. These ingredients can then be processed into different shapes, resulting in a wide variety of different materials with different properties.
Polymers
Polymers are long molecules. They are one of the basic components of all plastics.
Synthetic polymers
Synthetic polymers are polymers that are man-made. Most synthetic polymers are manufactured from petroleum.
Some examples of synthetic polymers include:
- polystyrene is the polymer found in styrofoam, used for everything from packing materials and insulation to drinking cups.
- polyvinyl chloride, widely known by its abbreviation PVC, is used in a lot of building material (and is well-known as being ubiquitous in piping).
These materials are generally not biodegradable, and because they are made from petroleum, once the basic materials for creating them are used up, we cannot make any more.
Biopolymers
Biopolymers are polymers that occur in nature. Carbohydrates and proteins, for example, are biopolymers. Many biopolymers are already being produced commercially on large scales, although they usually are not used for the production of plastics. Even if only a small percentage of the biopolymers already being produced were used in the production of plastics, it would significantly decrease our dependence on manufactured, non-renewable resources.
Some examples of biopolymers include:
- cellulose is the most plentiful carbohydrate in the world; 40 percent of all organic matter is cellulose!
- starch is found in corn (maize), potatoes, wheat, tapioca (cassava), and some other plants. Annual world production of starch is well over 70 billion pounds, with much of it being used for non-food purposes, like making paper, cardboard, textile sizing, and adhesives.
- collagen is the most abundant protein found in mammals. Gelatin is denatured collagen, and is used in sausage casings, capsules for drugs and vitamin preparations, and other miscellaneous industrial applications including photography.
- casein, commercially produced mainly from cow’s skimmed milk, is used in adhesives, binders, protective coatings, and other products.
- soy protein and zein (from corn) are abundant plant proteins. They are used for making adhesives and coatings for paper and cardboard.
- polyesters are produced by bacteria, and can be made commercially on large scales through fermentation processes. They are now being used in biomedical applications.
- A number of other natural materials can be made into polymers that are biodegradable. For example:
- lactic acid is now commercially produced on large scales through the fermentation of sugar feedstocks obtained from sugar beets or sugar cane, or from the conversion of starch from corn, potato peels, or other starch source. It can be polymerized to produce polylactic acid.
- triglycerides can also be polymerized. Triglycerides make up a large part of the storage lipids in animal and plant cells. Over sixteen billion pounds of vegetable oils are produced in the United States each year, mainly from soybean, flax, and rapeseed. Triglycerides are another promising raw material for producing plastics.
- These natural raw materials are abundant, renewable, and biodegradable, making them attractive feedstocks for bioplastics.
Plasticizers
A plasticizer is a substance that can be added to material to increase its workability, flexibility, or pliability. Plasticizers are one of basic ingredients of all plastics.
Additives
An additive is a substance that can be added to material to change its properties, usually to make the end product more desirable in some way. Additives are one of basic ingredients of all plastics.
The pure polymer resin by itself may not always have the physical properties needed in the final product, it may be strong but too brittle, flexible but too elastic, or flexible and elastic but just plain ugly. Just like the polymer material itself, additives come in different varieties: some can be found in the environment, while others are manufactured. The amounts and types of additives used in manufacturing plastics are another factor that influence how environmentally-friendly they are.
Green Plastics
Green Plastics, sometimes also called Bioplastics, are plastics that are biodegradable and are usually made mostly or entirely from renewable resources. Frequently there is also a focus on environmentally friendly processing. Green plastics are the focus of an emerging industry focused on making convenient living consistent with environmental stability.
Like all plastics, bioplastics are composed of a polymer, combined with plasticizers and additives, and processed using extrusion or thermosetting. What makes green plastics “green” is one or more of the following properties:
- they are biodegradable
- they are made from renewable ingredients
- they have environmentally friendly processing
- Because different compounds can satisfy some or all of these criteria to different degrees, there are different “degrees of green” in green plastics. To evaluate how “green” a plastic material is, you need to ask three questions:
- how quickly can the plastic be re-integrated into the environment after it is no longer being used?
- how quickly are the ingredients that go into making the plastic created in the environment?
- how much pollution or waste is created during the process of actually making the plastic?
Traditional plastics fail on all three of these points.
1. Biodegradability
Biodegradation is a process where something breaks down into simple compounds as a result of the action of microorganisms (like bacteria, fungi, or algae). The term biodegradation is actually a contraction, short for “biotic degradation.” Something is biodegradable if it can be broken down by this kind of process.
In order to say that something “biodegrades”, it therefore has to meet the following requirements:
- it has to break down (this is simply “degradation”)
- it’s molecules have to break down from complex molecules into simpler ones (this is “chemical degradation”)
- the breaking down of its molecules has to be accomplished by microorganisms.
- In order to prevent misinformation in advertising, standards organizations have made even more strict requirements for something to be labelled as biodegradable. In addition to the above list, something can only be labelled as “biodegradable” if:
- The biodegradation of the material has to be scientifically measurable. Since most biodegradation produces CO2 as a by-product, usually this is measured by the amount of CO2 produced.
- The biodegradation of the material has to be fast enough to have a significant effect in a reasonable amount of time. For example, the ISO standard requires 60% biodegradation within 180 days for a material to be called biodegradable; the Europenan Norm EN13432 is more strict, requiring 90% biodegradation within 90 days.
Types of Biodegradation
Because biodegradation requires microorganism to do something to a material, usually the material has to be broken up into smaller pieces first. As a result, most biodegradable materials become biodegradable after the action of another kind of degradation.
Hydro-biodegradable
Hydro-biodegradable materials are first broken down by interaction with water (a process called hydrolysis), and then are further broken down by microorganisms.
Photo-biodegradable
Photo-biodegradable materials are first broken down by interaction with sunlight (a process called photolysis), and then are further broken down by microorganisms.
Oxo-degradable
Some companies have been claiming that they have created an additive that can be added to traditional plastics to make them biodegradable. These products become what is called oxo-degradable, and sometimes is incorrectly identified as oxo-biodegradable.
Although this allows the plastic to return to the environment, these products are not biodegradable. Instead, the additive allows the plastic material to break down physically when exposed to water, into pieces small enough to be accidentally ingested by microbes. However, the microbes are not able to actually break this material down further. The end result is therefore a material that combines biomass with polymer residue. The plastic never decomposes as a result of interaction with the organisms. This process is therefore more accurately called “disintergration” rather than “biodegradation”.
For bioplastics to become practical, they must have properties that allow them to compete with the current plastics on the market: bioplastics must be able to be strong, resiliant, flexible, elastic, and above all, durable. It is the very durability of traditional plastics that has helped them in the marketplace, and has been a major goal of plastics research throughout the years. However, it is exactly this durability that now has people increasingly worried. Now that we wrap our sandwiches in bags that will still be around when the sandwich, and even the person who ate it, are long gone, many people are wondering: have we gone too far?
There is a lot of current research going on concerning methods of decomposition. There is also research on controlling the time-line of biodegradation. One goal of this research is to make a product that is programmed-degradable: in other words, a product that allows you to control when and how it degrades, while insuring that the product remains strong while it is still in use.
2. Renewability
A renewable resource is a natural resource that is created in the environment faster than it is used up by people. Many people think of “renewability” as a fixed trait: some things (like trees, grass, and wind) are renewable, while others (like oil and coal) are not. In fact, whether a resource is renewable depends on both how fast it is replenished and how fast people use it. As a result, some resources are more renewable than others, and some resources may or may not be renewable depending on how they are used.
Rate of Renewal
The rate of rewal (sometimes also called the “sustainable yield”) of a resource tells you how quickly it can be replenished by the environment.
Solar energy, tides, rainfall, and winds are considered perpetual resources for energy because they renew much faster than they could ever be used. (Can you imagine us “using up” the wind, so that we would have to wait until the earth made more?)
Living organisms provide the majority of resources that are generally considered “renewable”, because they generally renew themelves within a reasonable amount of time relative to how quickly they are use. Agricultural feedstocks and marine feedstocks are two major categories of living organism feedstocks. Within this category, some organisms renew faster than others: for example, it takes much longer to grow a new tree than it does to grow grass.
Most of the resources that are considered “non-renewable” are based on coal, oil, natural gas, and other substances that take so long for the environment to create that almost any use of these resources at all will cause them to be used up before any more is created. Petro-chemical feedstocks are feedstocks derived from petroleum principally for the manufacture of chemicals, synthetic rubber, and a variety of plastics.
Rate of Use
Imagine you live in a small village by a river. A turbine on the river spins, and it can generate enough electricity for the entire village every day. Clearly, their hydroelectric power is a completely renewable resource. However, as the size of the village grows, their energy use grows. If eventually the needs of the village far outstrip the energy that can be provided by the turbine, then the hydroelectric energy from the river is no longer a renewable resource for the village: the rate of use has exceeded the rate of replenishment.
The same issue exists for the use of plants. As long as our use of (for example) corn remains moderate compared to the amount of corn produced, corn is a renewable resource. However, if our use of corn increases dramatically without a corresponding increase in corn crop production, then corn will cease to be a renewable resource: we will use it all up, and we will either have to cease production until the corn renews itself or (worse) it will become extinct, so it will not replenish at all.
3. Processing
When making plastics, the initial mass of polymer, called resin, is processed into different shapes using a variety of methods, including: extrusion, injection molding, compression molding, transfer molding, and casting. Different processing techniques result in the wide variety of forms that plastic can take: ranging from thin films and elastic sheets, to resiliant panels and hard, solid three-dimensional shapes.
HISTORY OF BIOPLASTICS
The use of natural polymers is not entirely a new idea. In one form or another, green plastics have been around for a long time.
Early History
Natural resins-like amber, shellac, and gutta percha-have been mentioned throughout history, including during Roman times and the Middle Ages. Native Americans were developing and refining techniques for making ladles and spoons from animal horns long before there was any European contact. In Europe, molded horn jewelry and snuff boxes were popular in the eighteenth century.
The 1800’s
Significant commercialization of bioplastics only began in the middle of the nineteenth century… The American inventor, John Wesley Hyatt, Jr., was looking for a substitute for ivory in the manufacture of billiard balls, and in 1869 patented a cellulose derivative for coating non-ivory billiard balls. That attempt, however, was affected by the coating’s flammability; balls were occasionally ignited when lit cigars accidentally came into contact with them. Hyatt continued working on the project and soon developed celluloid, the first widely used plastic, now most widely known for its use in photographic and movie film.
The 1900’s
The history of plastics changed dramatically in the early 1900s, as petroleum emerged as a source of fuel and of chemicals. The early bioplastics were simply displaced by plastics made from synthetic polymers. World War II brought on a large increase in plastics production, a growth which continues to this day.
The 1920’s
In the 1920s Henry Ford experimented with using soybeans in the manufacture of automobiles. Ford was partly motivated by a desire to find non-food applications for agricultural surpluses, which existed then as they do now. Soy plastics were used for an increasing number of automobile parts, like steering wheels, interior trim, and dashboard panels. Finally Ford gave the go-ahead to produce a complete prototype “plastic car.” Ford, a master at generating publicity, exhibited the prototype with great fanfare in 1941, but by the end of the year was no longer publicizing the “plastic car,” probably for a variety of reasons. World War II played a role: armament work took precedent over almost everything else, and steel shortages limited all non-defense production. Today plastic automobile parts are common, but the use of plastics made from renewable raw materials got side-tracked.
The 1960’s
One well established bioplastic that has survived the growth of the synthetic plastics industry is cellophane, a sheet material derived from cellulose. Although production peaked in the 1960s it is still used in packaging for candy, cigarettes, and other articles.
The 2000’s and Beyond
Demand for materials like plastics is continually growing and will not be abated. Today, the plastics industry is an important component of our economy: The U.S. plastics industry includes over 20,000 facilities that produce or distribute materials or products, employ over 1.5 million workers, and ship over $300 billion in products each year.
The magnitude of the plastics industry, however, is itself a cause for concern. The pressures of increasing waste and diminishing resources have lead many to try to re-discover natural polymers and put them to use as materials for manufactor and industry. As a result, there is increasing interest in the promise of a new generation of green plastics.
| Download this lecture as PDF here |
Plant – Microbe symbioses
1. Many microbes (bacteria, fungi) have important symbioses with plants
2. Rhizosphere = thin layer of soil immediately attached to root hairs of plants. Typically contains 109 microbes/g of soil.
3. Many rhizosphere organisms are ectosymbionts, living outside the roots. Others are endosymbionts, living inside or penetrating into plant roots.
4. Many of these bacteria contribute Nitrogen fixation, obtain plant nutrients in return (see below for Rhizobium symbiosis).
Rhizobium-Legume symbioses
5. Plants of the legume family (soybeans, clover, alfalfa, beans, peas) can grow in soils lacking nitrogen compounds required by other plants. How?
6. These plants contain endosymbiotic Rhizobium bacteria that grow in root nodules. Rhizobia can fix atmospheric Nitrogen gas (N2)
N2 + 6[H] 2 NH3
7. The reaction requires total lack of oxygen and lots of energy as ATP. To bind oxygen and get rid of it, bacteria use protein called leghemoglobin, somewhat similar to animal hemoglobin. Globin part is encoded in plant genome, heme group is encoded in bacterial genome. Neither partner can fix nitrogen alone, only in symbiosis.

Hydrothermal vent Communities
- Occurs only near thermal springs on ocean floor, 2 miles or more below surface. Totally black, no sunlight penetrates below 600 feet.
- Associated with spreading centers of tectonic plates where hot magma close to surface causes area of floor to slowly drift apart.
- Seawater seeps down, mixes w/ minerals at high temperature comes back to ocean water in plumes at 270-380 deg. C. These are sometimes called black smokers since minerals precipitate as black cloud when in contact with cold sea water .
- Contains high levels of inorganics: Mn2+, H2, usually H2S; very low in organic matter
- Astonishing discovery: such regions are densely populated by a community of unusual animals: 2 m long tube worms, giant clams, mussels, white shrimp.
- What do they eat? Unlike earth’s surface, there is no source of light to stimulate phototrophs, they “eat” chemolithotrophic bacteria!
- Example: Inside the tube worms live huge colonies of bacterial endosymbionts. These are autotrophic chemolithotrophs, oxidizing sulfide to sulfate as their energy source. As bacteria grow, they provide carbon and nitrogen compounds for worms to feed on. Have not been cultivated outside of host, so little is known about details of the bacterium.
Ruminant Symbiosis
- Ruminants are the herbivorous mammals whose digestive tract contains four chambers. First chamber (largest) is the rumen, provides a place for bacteria to break down the fiber in the plants so the cow can use it for energy.Ý
- Includes cows, sheep, giraffes, buffalo, and elk.
- Ruminants eat grasses and other plant materials, but do not produce enzymes to digest cellulose, the primary plant metabolite.
- Instead, ruminants rely on huge microbial community in rumen to digest plant materials. Microbial densities can reach as high as 1012 microorganisms/ml, the highest density found anywhere in nature.
- Ruminants feed off fermentation waste products of microorganisms; mainly acetic acid, propionic acid, and butyric acid.
Gnotobiotic Animals
- Gnotobiotic = “known microbiota”; animal host is either entirely free of microbes (aka “germfree”, “axenic”) or has a microbiota whose identity is completely known.
- Animals in utero are germfree, but acquire resident bacteria within hours of birth.
- Relatively easy to produce germfree animals for birds. Sterilize shell, use sterile incubator, keep animals in an environment where all air, food, water is sterilized before entry.
- More difficult to establish germfree animals other than birds. Need cesarean section of pgrenant females, germfree isolation chambers where all air, food, water is sterilized before entry.
- Germfree animals generally are less healthy than animals with normal microbiota. Defects include:
- Greater vitamin requirements for K and B complex
- lower cardiac output
- much more susceptible to pathogens — normal microbiota colonize access sites, often compete successfully to prevent pathogens from binding to host tissues.much smaller infectious dose required to initiate an infection
Interrelationship between microorganisms: Beneficial and harmful relationships
Interrelationships in soils are of 3 types
- Plant microbe interaction
- Microbe – microbe interaction and
- Plant microbe – microbe interaction
a) Plant microbe interaction
It mainly constitutes the association of microorganism with plants little in a positive way or in a negative way. The positive approach is mainly the symbiotic relationships and the negative approach constituents mainly pathogen plant interactions.
b) Plant microbe – microbe interaction
Also called tripartite symbiosis
Eg: Alnus – Frankia –Mycorrhiza and Casbarina – Frankia – Mycorrhiza
Ceanothus roots, with Frankia vesicles

c) Microbe – microbe interaction
Microbial interaction in soil
Interrelationship between microorganisms: Beneficial and harmful relationship
Microorganisms live in the soil, not in the form of pure culture, but as complex populations. Each particle of soil contains more than one type of organisms. So, microbial ecosystem of soil is the sum of the biotic and the abiotic components of soil. Many of these organisms depend upon one another for direct and indirect nutrients. Some complete with one another for energy sources and for the elements and components used as nutrients. This results in the formation of numerous associations among the soil micro organic. The composition of the microflora of any habitat is governed by the biological equilibrium created by the associations and interactions of all individual found in the community.
The micro organic that inhabits the soil exhibited many different types of associations or interactions. Some of the associations are indifferent or neutral, some are beneficial type of interactions and others are detrimental or negative.
Beneficial / positive interactions
a. neutralism
b. Symbiosis / mutualism
c. Protoco-operation
d. Communalism
a) Neutralism
It is a type of neutral association, in two microorganisms behaves entirely independently or eg: Each could utilize different nutrients with out producing metabolic end products that are inhibitory. This might be transitory as the condition change in the environment, parituclary the availability of nutrients, the relationship might change.
Symbiosis / Mutualism
Mutualism is a form of symbiosis in which both organisms benefit. An example of mutualism is a clownfish and sea anemones. The clownfish gets protection, while the sea anemones become clean. This is mutualism, because both water animals benefit from having each other around


b) Proto co-operation
One type of mutualistic association is that involving the exchange of nutrients between two species, a phenomenon called syntrophism.
Many micro organic synthesize the vitamins and anaerobic acids in excess of their nutritional requirements. Others have a requirement of one or more of these nutrients. Hence certain combinations of species will grow together but not apart when nutrient levels are very low.
Nutritional proto co-operation has been demonstrated in cultures. Eg: In a medium deficient in nicotinic acid and biotin, neither Proteus vulgaris nor Bacillus polymyxa will multiply as the former (B) requires nicotinic acid and the latter biotin. In mixed culture, in the same medium however both grown since the partner bacterium synthesizes the missing vitamins.
c) Symbiosis
The living together of two or more organisms; microbial association
Symbiotic association is evident in soil among several groups of organisms algae and fungi in lichens, bacteria residing with in protozoa cells, bacteria and roots in the legume symbiosis, fungi and roots in mycoorhizae.
In lichens, the algae and fungi are in such an intimate physical and physiological relationship that the lichens they make are classified as distinct organism. The alga benefits in part are se of the protection afforded to it by the hyphae that envelop and protect it from environmental stresses. While, the fungi gains by making use of the CO2 fixed by its photosynthetic partner. Where BGA participants, the heterotraph benefits from the fixed N2.
Symbiotic relationship exists between micro and macro organisms. R-L associate N2 fixed is transferred to legume and organic which is transferred to the ® by CO2 metabolizing legume host.
Anabaena with heterocysts
d) Commensalisms
It is the type of beneficial association, in which only one species derives benefit while the other is unaffected. This occurs commonly in soil with respect to degradation of complex molecules like cellulose and lignin. One patter can attack a substrate not available to the second organism, but the decomposition results in the formation of products utilized by the second. The one which offer eg: (1) Many fungi able to degrade cellulose and yield glucose and organic acids. This can serve as a which source for many bacteria and fungi, which are non cellulolytic (2) The second type of commensal association arises from the need of many micro organic for growth factors. These compounds are synthesized by many micro organisms and their exertion permits the proliferation of nutritionally fastidious soil inhabitants.

III. Negative / harmful / deleterious interactions
Detrimental effects of one species on its neighbours are quite common in soil, and they are ditched by the decreases in abundance or metabolic activities of the susceptible organisms.
This include
a) Competition
b) Amensalism
c) Parasitism and predation
a. Competition
It is the rivalry for limiting nutrients or other common needs. In such situations the best adapted microbial species will predominate or infact, eliminate other species which are dependent upon the same limited nutrient substances.
Eg: Competition between strains derived from soil and those applied with legume seeds at the time of sowing. The better competitor involves the root hairs more frequently and it is responsible for a high % of nodules.

Sea Anemones compete for the territory in tide pools
b. Amensalism
It is a negative interaction, in which the release of products by one species is toxic to its neighbours. Antagonism is a type of ammensalism.
. Antagonism The killing, injury or inhibition of growth of one species of micro organisms by another or when one organism adversely affects the environmental of the other is refered as antagonism.
The killing, injury or inhibition of growth of one species of micro organisms by another or when one organism adversely affects the environmental of the other is refered as antagonism.
The toxic compounds are antibiotic. An antibiotic is a substance formed by one organic that in low concentrations inhibits the growth of another organism. Antibiotics are common among Streptomyces isolates, but numerous strains of Micromonespora and Nocardia are also active. The most common frequently encountered (B) synthesis antibiosis are species of Bacillus strains of Pseudomonas species of Peniciliu, Trichoderma, Aspgerillus, Fusarium are also excrete antibiotic substance.
Anitimicrobial compounds against (F) are present in the soil, which inhibit the germination of fungal spores. This phenomenon is termed as fungistasis. Cyanide is produced by certain (F) in concentrations toxic to other microorganisms, and algae elaborate fatty acids which exhibit and marked antibacterial activity other metabolic products that may result from microbial activity in soil, which are likely to be inhibiting to other species are CH4, sulfides and other volatile S compounds. B. t toxin to lepidopteran insect pestd
Myxobacteria (slime (B)) and streptomyces are antagonistic because they secrete potent lytic enzymes which destroy other cells by digesting their cell wall. The degraded cellular material as well as they released protoplasmic material, which serve as nutrients.
d. Predation
Direct attack of one organism on another predation is one of the most dramatic interrelationships among the micro organic in nature of the many microscopic inhabitants of soil, the bacteria stand out as particularly prove to the attach of predators. The most numerous predators on (B) are protozoans, which by feeding on the billions of (B) undisputedly affect their populations. Protozoans are a key factor in limiting the size of bacterial populations. Probably reducing the abundance of cells and serving to maintain a diverse community.
- Myxobacteria and cellular slime molds also affect by feeding directly on them
- Bacteria of the diverse genera are attacked by bacteriophages
- Bdellovibrio is ubiquitous, capable of attacking a number of bacterial genera.
- Parasitism is between two types of (B), or between different organisms of the same group (F, B, A).
- Creation of conditions by one organism which are unfavorable for the growth of another (change in pH).
- Production of specific substances by one organisms which are injurious to growth of other ( organic alcohols, quinones and antibiotics)
- Direct parasitism of one organism upon another- various effects of (F) upon (B), of (B) upon (F).

e. Parasitism
Is a form of symbiosis in which one organism benefits and the other is harmed. An example of parasitism is wasp’s eggs and caterpillar. When the eggs hatch into young wasps, these young wasps burrow into the body of the caterpillar. The young wasps feed on the caterpillar’s tissues. After a month or so, the young wasps chew their way out of the dying caterpillar’s body and spin cocoons. Afterwards, the young wasps become adult wasps. This is parasitism, because the caterpillar is harmed while the young wasps benefit from feeding on the caterpillar.


| Download this lecture as PDF here |
Introduction
In the twentieth century, the ever increase in the global human
population and industrialization led to the exploitation of natural resources. The increased usage of heavy metals
and its disposal in the ecosystems around the world and also in India is becoming an important ecological issue to be take into our consideration. This has indiscriminate release of heavy metals into the ecosystem has already posing air, water and soil pollution causing various
uncompromising, deleterious and fatal effects on humans and the
stability of the ecosystems.
Soil and water are the most important natural resources for all living beings including human beings for survival, which are recently becoming highly polluted. As a result of this, several living forms which are better suited to the polluted environment have out bursted causing ecological imbalance. Unlike soil, water body has its own ability to maintain its natural state through process called self purification. However when the discharge of pollutants is heavy, the process of self purification of water bodies is adversely affected and the water remains polluted.
Unlike organic contaminants,
heavy metals are not biologically degradable, and
therefore can persist in the environment for a long
duration. The term ‘heavy metal’ can be explained as 1) Relatively
abundant in the earth’s crust; 2) Reasonable extraction
and usage; 3) Having direct contact with people; and 4)
Toxic to humans. Heavy metals are the metals which have a specific gravity of more than 4 or more than 5 (Anonymous, 1964; Nieboer and Richardson, 1980). Some important heavy metals are Zinc (Zn), Chromium (Cr), Cadmium (Cd), Arsenic (As) etc.,
are the metals which have a specific gravity of more than 4 or more than 5 (Anonymous, 1964; Nieboer and Richardson, 1980). Some important heavy metals are Zinc (Zn), Chromium (Cr), Cadmium (Cd), Arsenic (As) etc.,
Bioremediation can be defined as any process that uses microorganisms, fungi, green plants or their enzymes to return the natural environment altered by contaminants to its original condition. Bioremediation may be employed to attack specific soil contaminants, such as degradation of chlorinated hydrocarbons by bacteria. An example of a more general approach is the cleanup of oil spills by the addition of nitrate and/or sulfate fertilisers to facilitate the decomposition of crude oil by indigenous or exogenous bacteria.
Mycoremediation is a form of bioremediation that uses fungi to reduce the level of contamination in a given environment. Fungi can release specific enzymes and acids that break down the major components of plant fiber. The accumulation of waste is directly proportional to population and as our population grows scientists will be under more pressure to find ways to eliminate contaminants from our environment. Bioremediation microbes will continue to be used in an attempt to return our polluted environments to their original state. Corning, Barnstead, and BD supply products used by those researching bioremediation.
Bioremediation technologies can be generally classified as in situ or ex situ. In situ bioremediation involves treating the contaminated material at the site while ex situ involves the removal of the contaminated material to be treated elsewhere. Some examples of bioremediation technologies are bioventing, landfarming, bioreactor, Composting, bioaugumentation, rhizofiltration, and biostimulation.

BIOREMEDIATION STRATEGIES
Different techniques are employed depending on the degree of saturation and aeration of an area. In situ techniques are defined as those that are applied to soil and groundwater at the site with minimal disturbance. Ex situ techniques are those that are applied to soil and groundwater at the site which has been removed from the site via excavation (soil) or pumping (water). Bioaugmentation techniques involve the addition of microorganisms with the ability to degrade pollutants.
In situ bioremediation
These techniques are generally the most desirable options due to lower cost and less disturbance since they provide the treatment in place avoiding excavation and transport of contaminants. In situtreatment is limited by the depth of the soil that can be effectively treated. In many soils effective oxygen diffusion for desirable rates of bioremediation extend to a range of only a few centimeters to about 30 cm into the soil, although depths of 60 cm and greater have been effectively treated in some cases. The most important land treatments are:
Incineration Excavation

Bioventing
Is the most common in situ treatment and involves supplying air and nutrients through wells to contaminated soil to stimulate the indigenous bacteria. Bioventing employs low airflow rates and provides only the amount of oxygen necessary for the biodegradation while minimizing volatilization and release of contaminants to the atmosphere. It works for simple hydrocarbons and can be used where the contamination is deep under the surface. 
In situ biodegradation
Involves supplying oxygen and nutrients by circulating aqueous solutions through contaminated soils to stimulate naturally occurring bacteria to degrade organic contaminants. It can be used for soil and groundwater. Generally, this technique includes conditions such as the infiltration of water-containing nutrients and oxygen or other electron acceptors for groundwater treatment.
Biosparging.
Biosparging involves the injection of air under pressure below the water table to increase groundwater oxygen concentrations and enhance the rate of biological degradation of contaminants by naturally occurring bacteria. Biosparging increases the mixing in the saturated zone and there- by increases the contact between soil and groundwater. The ease and low cost of installing small-diam- eter air injection points allows considerable flexibility in the design and construction of the system.
Bioaugmentation.
Bioremediation frequently involves the addition of microorganisms indigenous or exogenous to the contaminated sites. Two factors limit the use of added microbial cultures in a land treatment unit: 1) nonindigenous cultures rarely compete well enough with an indigenous population to develop and sustain useful population levels and 2) most soils with long-term exposure to biodegradable waste have indigenous microorganisms that are effective degrades if the land treatment unit is well managed.
Ex situ bioremediation
These techniques involve the excavation or removal of contaminated soil from ground.
Landfarming Is a simple technique in which contaminated soil is excavated and spread over a prepared bed and periodically tilled until pollutants are degraded. The goal is to stimulate indigenous biodegradative microorganisms and facilitate their aerobic degradation of contaminants. In general, the practice is limited to the treatment of superficial 10–35 cm of soil. Since landfarming has the potential to reduce monitoring and maintenance costs, as well as clean-up liabilities, it has received much attention as a disposal alternative.
Is a simple technique in which contaminated soil is excavated and spread over a prepared bed and periodically tilled until pollutants are degraded. The goal is to stimulate indigenous biodegradative microorganisms and facilitate their aerobic degradation of contaminants. In general, the practice is limited to the treatment of superficial 10–35 cm of soil. Since landfarming has the potential to reduce monitoring and maintenance costs, as well as clean-up liabilities, it has received much attention as a disposal alternative.
Composting
Is a technique that involves combining contaminated soil with nonhazardous organic amendants such as manure or agricultural wastes. The presence of these organic materials supports the development of a rich microbial population and elevated temperature characteristic of composting.
Biopile
Are a hybrid of landfarming and composting. Essentially, engineered cells are constructed as aerated composted piles. Typically used for treatment of surface contamination with petro- leum hydrocarbons they are a refined version of landfarming that tend to control physical losses of the contaminants by leaching and volatilization. Biopiles provide a favorable environment for indigenous aerobic and anaerobic microorganisms.

Bioreactors
Slurry reactors or aqueous reactors are used for ex situ treatment of contaminated soil and water pumped up from a contaminated plume. Bioremediation in reactors involves the processing of contaminated solid material (soil, sediment, sludge) or water through an engineered containment system. A slurry bioreactor may be defined as a containment vessel and apparatus used to create a three-phase (solid, liquid, and gas) mixing condition to increase the bioremediation rate of soil- bound and water-soluble pollutants as a water slurry of the contaminated soil and biomass (usually indigenous microorganisms) capable of degrading target contaminants. In general, the rate and extent of biodegradation are greater in a bioreactor system than in situ or in solid-phase systems because the contained environment is more manageable and hence more controllable and predictable. Despite the advantages of reactor systems, there are some disadvantages. The contaminated soil requires pre treatment (e.g., excavation) or alternatively the contaminant can be stripped from the soil via soil washing or physical extraction (e.g., vacuum extraction) before being placed in a bioreactor.

Advantages of bioremediation
• Bioremediation is a natural process and is therefore perceived by the public as an acceptable waste treatment process for contaminated material such as soil. Microbes able to degrade the contaminant increase in numbers when the contaminant is present; when the contaminant is degraded, the biodegradative population declines. The residues for the treatment are usually harmless products and include carbon dioxide, water, and cell biomass.
• Theoretically, bioremediation is useful for the complete destruction of a wide variety of contaminants. Many compounds that are legally considered to be hazardous can be transformed to harmless products. This eliminates the chance of future liability associated with treatment and disposal of contaminated material.
• Instead of transferring contaminants from one environmental medium to another, for example, from land to water or air, the complete destruction of target pollutants is possible.
• Bioremediation can often be carried out on site, often without causing a major disruption of nor- mal activities. This also eliminates the need to transport quantities of waste off site and the poten- tial threats to human health and the environment that can arise during transportation.
• Bioremediation can prove less expensive than other technologies that are used for clean-up of hazardous waste.
Disadvantages of bioremediation
• Bioremediation is limited to those compounds that are biodegradable. Not all compounds are susceptible to rapid and complete degradation.
• There are some concerns that the products of biodegradation may be more persistent or toxic than the parent compound.
• Biological processes are often highly specific. Important site factors required for success include the presence of metabolically capable microbial populations, suitable environmental growth conditions, and appropriate levels of nutrients and contaminants.
• It is difficult to extrapolate from bench and pilot-scale studies to full-scale field operations.
• Research is needed to develop and engineer bioremediation technologies that are appropriate for sites with complex mixtures of contaminants that are not evenly dispersed in the environment.
Contaminants may be present as solids, liquids, and gases.
• Bioremediation often takes longer than other treatment options, such as excavation and removal
of soil or incineration.
• Regulatory uncertainty remains regarding acceptable performance criteria for bioremediation.
There is no accepted definition of “clean”, evaluating performance of bioremediation is difficult, and there are no acceptable endpoints for bioremediation treatments.
| Download this lecture as PDF here |
Biosensor is an analytical device for the detection of an analyte that combines a biological component with a physicochemical detector component
It consists of 3 parts:
- The sensitive biological element (biological material (eg. tissue, microorganisms, organelles, cell receptors, enzymes, antibodies, nucleic acids, etc), a biologically derived material or biomimic) the sensitive elements can be created by biological engineering.
- The transducer or the detector element (works in a physicochemical way; optical, piezoelectric, electrochemical, etc.) that transforms the signal resulting from the interaction of the analyte with the biological element into another signal (i.e., transducers) that can be more easily measured and quantified;
- Associated electronics or signal processors that are primarily responsible for the display of the results in a user-friendly way. This sometimes accounts for the most expensive part of the sensor device, however it is possible to generate a user friendly display that includes transducer and sensitive element (see Holographic Sensor).
A common example of a commercial biosensor is the blood glucose biosensor, which uses the enzyme glucose oxidase to break blood glucose down. In doing so it first oxidizes glucose and uses two electrons to reduce the FAD (a component of the enzyme) to FADH2. This in turn is oxidized by the electrode (accepting two electrons from the electrode) in a number of steps. The resulting current is a measure of the concentration of glucose. In this case, the electrode is the transducer and the enzyme is the biologically active component.
Recently, arrays of many different detector molecules have been applied in so called electronic nose devices; where the pattern of response from the detectors is used to fingerprint substance. Current commercial electronic noses, however, do not use biological elements.
Principles of Detection
Analytical chemistry plays an important role in food quality parameters because almost every sector of industry and public service relies on quality control. A food quality biosensor is a device, which can respond to some property or properties of food and transform the response(s) into a detectable signal, often an electric signal. This signal may provide direct information about the quality factor(s) to be measured or may have a known relation to the quality factor. There are various kinds of biosensors most of which work on the principle of one of the following:
Electrochemical Biosensors
Electrochemical biosensors are based on monitoring electroactive species that are either produced or consumed by the action of the biological components (e.g., enzymes and cells). Transduction of the produced signal can be performed using one of several methods under two broad headings:
- Potentiometric Biosensors
- Amperometric Biosensors
Potentiometric Biosensors
These are based on monitoring the potential of a system at a working electrode, with respect to an accurate reference electrode, under conditions of essentially zero current flow. In process, potentiometric measurements are related to the analyte activity (of a target species) in the test sample. Potentiometric biosensors can operate over a wide range (usually several orders of magnitude) of concentrations. The use of potentiometric biosensors for food quality analysis has not been as widely reported as for amperometric sensors. However, some of the examples where this approach has been used for food quality analysis include estimating monophenolase activity in apple juice, determining the concentration of sucrose in soft drinks, measuring isocitrate concentrations in fruit juices, and determining urea levels in milk.
Amperometric Biosensors
The use of amperometric biosensors in signal transduction has proved to be the most widely reported using an electrochemical approach. Both “one-shot” (disposable) sensors and on-line (multi measurement) devices are commercially available, monitoring a wide range of target analytes. In contrast to potentiometric devices, the principle operation of amperometric biosensors is defined by a constant potential applied between a working and a reference electrode. The applied potential results in redox reactions, causing a net current to flow. The magnitude of this current is proportional to the concentration of electro active species present in test solution and both cathodic (reducing) and anodic (oxidizing) reactions can be monitored amperometrically. Most of the amperometric biosensors described use enzymes as the biorecognition element. Typically, oxidase and dehydrogenase enzymes have been the most frequently exploited catalysts used for these biosensor formats.
Calorimetric Biosensors
Most of the biochemical reactions are accompanied by either heat absorption or production. Sensors based on calorimetric transduction are designed to detect heat generated or consumed during a biological reaction; by using sensitive heat detection devices. Various biosensors for specific target analytes have been constructed. In the field of food quality analysis, uses of such biosensors to detect metabolites have been described.
Optical Biosensors
These sensors are based on measuring responses to illumination or to light emission. Optical biosensors can employ a number of techniques to detect the presence of a target analyte and are based on well-founded methods including chemiluminescence, fluorescence, light absorbance, phosphoresence, photothermal techniques, surface plasmon resonance (SPR), light polarization and rotation, and total internal reflectance. For example the use of this technique has been demonstrated to detect the presence of allergens, in particular peanuts, during food production.
Acoustic Biosensors
Piezoelectric quartz crystals can be affected by a change of mass at the crystal surface; this phenomenon has been successfully exploited and used to develop acoustic biosensors. For practical applications, the surface of the crystal can be modified with recognition elements (e.g., antibodies) that can bind specifically to a target analyte.
Immunosensors
Immunosensors are based on exploiting the specific interaction of antibodies with antigens. Typically, immunoassays (such as the enzyme-linked immunosorbent assay technique) employ a label (e.g., enzyme, antibody, fluorescent marker) to detect the immunological reaction. The use of biosensor platforms, linked to an immunoassay format, offers a route to rapid and accurate quantitative measurements of target analytes.
Applications of Biosensors
There are many potential applications of biosensors of various types. The main requirements for a biosensor approach to be valuable in terms of research and commercial applications are the identification of a target molecule, availability of a suitable biological recognition element, and the potential for disposable portable detection systems to be preferred to sensitive laboratory-based techniques in some situations. Some examples are given below:
- Glucose monitoring in diabetes patients ←historical market driver
- Other medical health related targets
- Environmental applications e.g. the detection of pesticides and river water contaminants
- Remote sensing of airborne bacteria e.g. in counter-bioterrorist activities
- Detection of pathogens
- Determining levels of toxic substances before and after bioremediation
- Detection and determining of organophosphate
- Routine analytical measurement of folic acid, biotin, vitamin B12 and pantothenic acid as an alternative to microbiological assay
- Determination of drug residues in food, such as antibiotics and growth promoters, particularly meat and honey.
- Drug discovery and evaluation of biological activity of new compounds.
- Protein engineering in biosensors
- Detection of toxic metabolites such as mycotoxins
Utility Biosensors for applications in Agriculture in Food/ Fruit Quality Control
Quality control is the essential part of a food industry and efficient quality assurance is becoming increasingly important. Consumers expect good quality and healthy food at a given price; with good shelf life and high safety while food inspections require good manufacturing practices, safety, labelling and compliance with the regulations. Further, food producers are increasingly asking for efficient control methods, in particular through on-line or at-line quality sensors. Their main aim is to satisfy the consumer and regulatory requirements and to improve the production feasibility, quality sorting, automation and reduction of production cost and production time subsequently.
Biochemical Composition of Fruits
The quality of soft fruit, in terms of taste, nutrition and consumers acceptance, is fundamentally based on the biochemical composition of the fruit. In soft fruits (viz. blackcurrant and strawberry) sugar: acid ratios can be used as an important index of fruit maturity and act as a determinant of overall fruit. However, sugar: acid ratios are infrequently used due to a requirement for specific instrumentation and semi-skilled analytical scientists. Today we need a simple and low-cost alternative, which would significantly enhance both the number and extent of tests carried out.
Fruit Maturity, Ripening and Quality Relationships
Fruit maturity at harvest is the most important factor that determines shelf life and final fruit quality. If harvested immature then fruits are more subject to shriveling and mechanical damage, and are of inferior quality when ripe, whereas overripe fruits are liable to become soft and mealy with bland flavour soon after harvest. Therefore, fruits harvested either too early or too late in their season are more susceptible to post harvest physiological disorders than fruits harvested at proper maturity.
Fruits can be divided into two groups:
- Fruit that are incapable of enduring their ripening process once picked from the plant like berries, cheery, citrus fruits, grapes, lychee, pineapple, pomegranate, and tamarillo.
- Fruits that can be harvested mature and ripped off the plant like apple, apricot, avocado, banana, cherimoya, guava, kiwifruit, mango, nectarine, papaya, passion fruit, pear, peach, persimmon, plum, quince, sapodilla, sapota.
Volatile compounds are responsible for the characteristic aroma of fruits and are present in extremely small quantities (<100< g/g fresh wt.). The major volatile formed is ethylene. Scientists are trying to develop portable instruments with sensors that detect volatile production by fruits and hence detecting maturity and quality. Other strategies include the removal of a very small amount of fruit tissue and measurement of total sugar or organic acid content.
Major organic acids in fruits
Organic acids function in growth, maturation, senescence, color, and antimicrobial activity of fruits. The low pH of fruits is due to the three most common organic acids present in fruits citric acid, malic acid, and tartaric acid. The total amount of acid in fruits varies widely, from about 0.2% in pear juice to 0.8% in limejuice. The amount and type of acid present in fruits determine the fresh taste of fruits and also affects the shelf life.
Organic Acid as an Indicator of Fruit Maturity
Organic acids directly play an important role in the growth, maturation and acidity of the fruit, and also affect the shelf life of the fruit by influencing the growth of microorganisms. The citric, malic, oxalic, and tartaric acids ranging from 0.1 to 30 g/L were found in orange, grape, and apple juices. There is a considerable difference in the organic acid content found in various types and brands of fruit juice. For example, Minute Maid contains higher levels of oxalic and citric acids when compared to all other orange juices tested. Grape concentrate was found to have lower amount of malic acid than other grape juice, while freshly squeezed grape juice contains higher amount of tartaric acid. Brae burn apples contained the highest amount of citric acid in apples; however Granny Smith apples were the overall most acidic apples tested.
Successful Examples of Organic Acid Biosensors Developed Pyruvic Acid
Onion flavour is principally directed by the perception of pungency. A disposable prototype electrochemical screen-printed (carbon-based) biosensor (C2030519D5, GEM Ltd., Gwent, UK) was constructed using pyruvate dehydrogenase immobilized on mediated Meldolas Blue electrodes and a combined Ag/AgCl reference/counter electrode, both screen-printed onto a PVC substrate to determine pungency in onions (Allium cepa L.). Electrochemical measurements were carried out using a Palm Sense potentiostat (Palm Instruments BV, The Netherlands). The biosensor developed was able to differentiate between mild and pungent bulbs with pyruvate concentrations ranging between 4 and 8 mM in freshly extracted juices. Electrochemical measurements were carried out at +50 mV at 21°C.
Glucose Biosensors
Most of the glucose biosensors developed are based on immobilized glucose oxidase. In many cases, glucose oxidase has been associated with mediators so as to bring down the high working potential required for hydrogen peroxide breakdown. The α-D glucose sensor developed was also based on glucose oxidase, at the working potential of -350 mV vs. Ag/AgCl, hydrogen peroxide was catalytically oxidized at a rhodinised carbon electrode (White et al, 1994). A novel and simple method which do not involve enzyme or monomer modifications, for the coimmobilization of ferrocene and GOx in a poly(pyrrole) matrix for use as glucose biosensor was developed (Foulds and Lowe, 1988). In spite of the low conductivity of the polypyrrole film formed, the biosensor’s performance was better than that of other devices reported due to redox mediation of ferrocene that lowers the working potential to 0.4 V. The characterization of the polymer prepared from an ethanolic suspension demonstrated the presence of alcohol interferes in the polymerization kinetics (Pablo et al., 2001). However, this played a beneficial role in efficient immobilization of both, the enzyme and the ferrocene, in a very thin electroactive film. This fact improved the biosensor’s time response, avoiding mass transport effects. A new type of disposable amperometric biosensor was devised by screen-printing thick-film electrodes directly on a porous nitrocellulose (NC) strip. A glucose biosensor based on hydrogen peroxide detection was constructed by immobilizing glucose oxidase (GOx) on the NC electrode strip and by formulating a strong oxidation layer (i.e., PbO2) at the sample loading area, placed below the GOx reaction band. The screen-printed PbO2 paste serves as a sample pretreatment layer that removes interference by its strong oxidizingability. Samples applied were carried chromatographically, via the PbO2 paste, to the GOx layer, and glucose was catalyzed to liberate hydrogen peroxide, which was then detected at the electrode surface. The proposed NC/ PbO2 strip sensor is shown to be virtually insusceptible to interfering species such as acetaminophen and ascorbic and uric acids and to exhibit good performance, in terms of the sensor to sensor reproducibility. The characterization of metal-decorated CNTs was done using X-ray diffraction analysis, transmission electron microscopy (TEM), high-resolution TEM, scanning electron microscopy, and energy-dispersive X-ray analysis. Amperometric biosensor fabricated by depositing GOD over Nafion-solubilized Au-MWNT electrode retained its biocatalytic activity and obtained fast and sensitive glucose quantification. The fabricated GOD/Au- MWNT/Nafion electrode has a good glucose biosensing potential, and it displayed a linear response up to 22 mM glucose and a detection limit of 20 ìM method.

Sucrose Biosensor
Sucrose is an essential part of any fruit, so estimating the concentration of sucrose at different maturity levels could help in identifying the ripening parameters of fruits. Therefore, with regard to sucrose detection, electrodes made up of invertase, mutarotase and glucose oxidase and mediated tri-enzyme electrode based on sucrose phosphorylase and electrocatalytic oxidation of NADH, have been used. Because real samples contain both glucose and sucrose, sucrose sensors have been operated in tandem with glucose oxidase sensors. The sucrose sensor developed was based on the invertase, mutarotase and glucose oxidase reaction scheme and the sucrose level was calculated with respect to the net glucose sensors.
Ascorbic Acid Sensor
Ascorbic acid has been measured both by direct electrochemical oxidation and by enzymatic methods using ascorbate oxidase. In the first case, an ascorbate oxidase electrode was used to measure the signal generated by other electroactive interferents in the analyte. The second method was based on the measurement of oxygen consumed during the enzyme-catalysed oxidation of ascorbic acid using Clark Electrode.
Lactic Acid Biosensor
The level of lactic acid in blood is used in clinical diagnostics of hypoxia, lactic acidosis, some acute heart diseases and drug toxicity tests. Reliable blood lactate measurements would also be of interest in sports medicine. Lactate can be measured based on the reaction using NAD+ dependent lactate dehydrogenase and ferricyanide. The concentration of dissolved L-lactate was determined in tomato paste and baby food samples using a SIRE-based (sensors based on injection of the recognition element) biosensor. The evaluation principle was based on the injection of small amount of enzyme into an internal delivery flow system and held in direct spatial contact with the amperometric transducer by the use of a semipermeable membrane. All the measurements were based upon the reversible enzymatic conversion of L lactate to pyruvate and hydrogen peroxide by lactate oxidase. The L-lactate concentrations of the tomato paste and baby food were calculated to be 1.02 (0.02 mM) and 2.51 (0.10 mM), respectively, using the standard addition method.
Phenolic Compounds
Phenolic compounds are widespread in nature, and they play a significant role in living organisms. They are used in medicine and industries, including wood processing and pesticide production. Most of the phenolic derivative compounds are highly toxic, and their determination in low concentrations is the significant problem. Scientists are developing various procedures for determining phenols with biosensors.
A biosensor based on crude seed hull enzyme extracts has been prepared for monitoring phenol and hydrogen peroxide. The biosensor has confirmed very promising results as a successful instrument to monitor both hydrogen peroxide and phenol. It is an inexpensive biosensor that could be operated for up to 3 weeks with rapid response and stability parameters. In conditions of response to phenol detection, the developed SBP biosensor was found less sensitive than other previously reported biosensors based on purified SBP or HRP or on crude extracts of sweet potato, which have detection limits in the micromolar range for phenols. The foremost reason for this was the low activity of the enzyme extracts. Further work on the improvement of biosensor sensitivity and applications for the detection of chlorophenols and other substituted phenols are in progress.
The amperometric biosensor described glucose oxidase and polyphenol oxidase carbon paste electrodes prepared via a new strategy of carbon paste modification based on the in situ electropolymerizaton of pyrrole monomer previously mixed within the paste. Such alteration induced a better electrical percolation of the carbon structure and enhanced the enzyme entrapment within the electrode material. Therefore, attractive potentialities offered by a biocomposite electrode based on PPO for the detection of flavonols have been demonstrated to control the phenolic levels in beer samples.
Benzoic Acid
An amperometric benzoic acid-sensing inhibitor biosensor was prepared by immobilizing mushroom (Agaricus bisporus) tissue homogenate on a Clarktype oxygen electrode. The effects of the quantity of mushroom tissue homogenate, the quantity of gelatin and the effect of the cross-linking agent glutaraldehyde percent on the biosensor were deliberated. The most favourable concentration of phenol used as substrate was 200 mM. The biosensor responded linearly to benzoic acid in a concentration range of 25–100 mM and Standard deviation (s.d.) was found to be ±0.49 μM for 7 successive determinations at a concentration of 75 μM. The inhibitor
biosensor based on mushroom tissue homogenate was applied for the determination of benzoic acid in fizzy lemonade, some fruits and groundwater samples. A good concord was shown when the results were compared to those obtained using AOAC
method.
Fructose
A superior amperometric biosensor based on a solid binding matrix (SBM) composite transducer has been used for the determination of d-fructose in various food samples. The enzyme, d-fructose dehydrogenase (EC 1.1.99.11), was incorporated directly into a solid composite transducer containing both 2-hexadecanone as SBM and chemically modified graphite. The current variation caused by the presence of d-fructose was calculated amperometrically using Hexacyanoferrate (iii) as a redox mediator. The amperometric signals generated were fast, reproducible and linearly proportional to d-fructose concentrations in the range 50×10-6 – 10×10-3mol l-1, with a correlation coefficient of 0.999. A set of measurements at +0.20 V versus SCE for 2×10-3 mol l-1 D-fructose yielded a relative standard deviation for the steady-state current of 2.11%. The biosensor selectivity against anionic interferents such as Lascorbate was enhanced by the use of chemically modified graphite by a mild oxidation step. The biosensor was found stable for 6 months and the assay of D-fructose by this electrode was not affected by the presence of sugars or other interferents commonly found in food samples.
ENVIRONMENTAL APPLICATIONS
Toxicity
In environmental pollution monitoring, it is becoming a general opinion that chemical analysis by itself does not provide sufficient information to assess the ecological risk of polluted waters and wastewaters. In the European Union, along with more stringent demands for water treatment (Council Directive 91/271/EEC), industrial and urban wastewater effluents shall reach certain limits of nontoxicity before the effluent can be discharged into the environment. Thus, much effort has been made during the last years to develop and use different bioassays and biosensors for toxicity evaluation of water samples. Whole organisms are used to measure the potential biological impact (toxicity) of a water or soil sample. That is the case of the toxicity assays Microtox® (Azure, Bucks, UK), or ToxAlert® (Merck, Darmstadt, Germany). These systems are based on the use of luminescent bacteria, Vibrio fischeri, to measure toxicity from environmental samples. Bacterial bioluminescence has proved to be a convenient measure of cellular metabolism and, consequently, a reliable sensor for measuring the presence of toxic chemicals in aquatic samples. Some bioassay methods are integrated now in biosensors such as the Cellsense®, which is an amperometric sensor that incorporates Escherichia coli bacterial cells for rapid ecotoxicity analysis. It uses ferricyanine, a soluble electron mediator, to divert electrons from the respiratory system of the immobilized bacteria of a suitable carbon electrode. The resulting current is, thus, a measure of bacterial respiratory activity, and the perturbation by pollutants can be detected as a change in the magnitude of the current. Cellsense has been applied to investigate the toxicity of 3,5-dichlorophenol and other phenols in wastewater, for the determination of nonionic surfactants and benzene sulfonate compounds, for the analysis of wastewater treatment works (WWTW) influent and effluent, and for the toxicity testing of wastewaters and sewage sludge. Moreover, Cellsense has been proposed as one of the newer rapid toxicity assessment methods within the direct toxicity assessment (DTA) demonstration program of the UK Environmental Agency. Most environmental biosensors have focused on bacterial systems while eukariotic biosensors are rare; even more rare is the use of mammalian cells. The mammalian cell, which is more complex than bacteria, can give a more sensitive response when compared to bacteria while also responding to the estrogenic effects of chemicals. A recombinant fluorescent Chinese Hamster Ovary cell line, utilizing a fluorescent reporter system, was used to monitor various toxicants, especially endocrine-disrupting compounds (EDCs), in diverse aqueous environments. EDCs have been also analyzed with a multichannel two-stage mini-bioreactor system using a genetically engineered bioluminescent bacteria. The toxicity of various samples spiked with known endocrine-disrupting chemicals, and phenol was investigated.
CONCLUSIONS
Despite the huge potential of biosensors, and the ever-increasing number of biosensors developed, commercially available biosensors are being applied to a restricted area of the potential market. In general, biosensors for environmental analysis have several limitations: sensitivity, response time, and lifetime, which should be improved for them to become a competitive analytical tool. The areas of development that are expected to have an impact in biosensor technology are: immobilization techniques, nanotechnology, miniaturization, and multisensor array determinations. However, a crucial aspect may be the production of new sensing elements easy to synthesize and with the capability to broaden the spectra of selectivities that can be reached by a biosensor. At present, the preparation and production in large scales of biomolecules such as enzymes or antibodies need an investment of time and knowledge. Synthetic peptides and MIPs are contemplated as promising alternatives overcoming the above-mentioned limitations. Unfortunately, the affinity accomplished by these synthetic receptors is still several orders of magnitude below that of the antibodies. Improvement in the affinity, specificity, and mass production of the molecular recognition components may ultimately dictate the success or failure of detection technologies. The possibility of tailor binding molecules with predefined properties, such as selectivity, affinity, and stability, is one of the major aims for biotechnology. The development of advanced receptors will allow the analysis of complex real samples and in situ measurements resolving the responses from the analyte and from nonspecific background effects. Since scientific attention is currently being given to biotechnology, as this review has pointed out, the development of improved molecular recognition elements will be followed by a corresponding enhancement of the biosensor features. From the above viewpoint, it is clear that the future of biosensors will rely on the success of emerging sophisticated micro and nanotechnologies, biochemistry, chemistry, thin-film physics, and electronics. To reach this goal, an important investment in research, expertise, and the necessary facilities is needed. However, as the world becomes more concerned about the impact that environmental contamination may cause on public health and the ecosystem, the demand for rapid detecting biosensors will only increase. Biosensors still need to achieve the confidence of potential users, having in mind that the commercialization of new devices will always be the best indicator of the success of a biosensor technology. The analysis of complex matrices and of analytes difficult to determine by the actual analytical procedures (i.e., highly polar compounds), are progressively being approached by biosensors. However, there is still a lack of alternative biosensing systems for an important bunch of emerging contaminants such as bisphenol A, phtalates, and polybrominated compounds (used as flame retardants), veterinary and human medicines and personal care products (nutraceuticals, synthetic fragances, sun screen agents, etc.)
| Download this lecture as PDF here |
The term Industrial Microbiology refers to the use of microorganisms for industrial purposes. Such things as anticoagulants, antidepressants, vasodilators, herbicides, insecticides, plant hormones, enzymes, and vitamins have been isolated from microorganisms or produced in large quantities by genetically engineering the organisms with foreign genes. In commercial industrial plants, microorganisms are widely used to produce numerous organic materials that have far-reaching value and application.
Antibiotic production
These are defined as substances produced by some micro-organisms which are in some way lethal to other micro-organisms. It is thought that these substances give the organisms that produce them (usually moulds or actinomycetes – which grow slowly) some sort of advantage in competition with other micro-organisms (usually bacteria – which grow fast) in the same habitat. However, their great medical advantage in healing infections is that the purified forms of antibiotics are more or less harmless to most humans. This means that they must act on some aspect of of the growth of micro-organisms which differs from ordinary mammalian cells. There are in fact several versions of Penicillin, variations on a common formula, produced by different strains of Penicillium, or using different culture media and methods.
The Production of Antibiotics has been widespread since the pioneering efforts of Florey and Chain in 1938. The importance of antibiotics to medicine has led to much research into their discovery and production.
Identifying Useful Antibiotics
![]()
An agar plate streaked with microorganisms.
Despite the wide variety of known antibiotics, less than 1% of antimicrobial agents have medical or commercial value. For example, whereas penicillin has a high therapeutic index as it does not generally affect human cells, this is not so for many antibiotics. Other antibiotics simply lack advantage over those already in use, or have no other practical applications.
Useful antibiotics are often discovered using a screening process. To conduct such a screen, isolates of many different microorganisms are cultured and then tested for production of diffusible products that inhibit the growth of test organisms. Most antibiotics identified in such a screen are already known and must therefore be disregarded. The remainder must be tested for their selective toxicities and therapeutic activities, and the best candidates can be examined and possibly modified.
A more modern version of this approach is a rational design program. This involves screening directed towards finding new natural products that inhibit a specific target, such as an enzyme only found in the target pathogen, rather than tests to show general inhibition of a culture.
Industrial Production Techniques
Antibiotics are produced industrially by a process of fermentation, where the source microorganism is grown in large containers (100,000–150,000 liters or more) containing a liquid growth medium. Oxygen concentration, temperature, pH and nutrient levels must be optimal, and are closely monitored and adjusted if necessary. As antibiotics are secondary metabolites, the population size must be controlled very carefully to ensure that maximum yield is obtained before the cells die. Once the process is complete, the antibiotic must be extracted and purified to a crystalline product. This is simpler to achieve if the antibiotic is soluble in organic solvent. Otherwise it must first be removed by ion exchange, adsorption or chemical precipitation.
Strains Used For Production
Microorganisms used in fermentation are rarely identical to the wild type. This is because species are often genetically modified to yield the maximum amounts of antibiotics. Mutation is often used, and is encouraged by introducing mutagens such as ultraviolet radiation, x-rays or certain chemicals. Selection and further reproduction of the higher yielding strains over many generations can raise yields by 20-fold or more. Another technique used to increase yields is gene amplification, where copies of genes coding for enzymes involved in the antibiotic production can be inserted back into a cell, via vectors such as plasmids. This process must be closely linked with retesting of antibiotic production and effectiveness.
WINE PRODUCTION
Winemaking, or Vinification, is the production of wine, starting with selection of the grapes or other produce and ending with bottling the finished wine. Although most wine is made from grapes, it may also be made from other fruit or non-toxic plant material. Mead is a wine that is made with honey being the primary ingredient after water.
Winemaking can be divided into two general categories: still wine production (without carbonation) and sparkling wine production (with carbonation).
The science of wine and winemaking is known as oenology (in American English, enology).
PROCESS
After the harvest, the grapes are taken into a winery and prepared for primary ferment, at this stage red wine making diverges from white wine making. Red wine is made from the must (pulp) of red or black grapes that undergo fermentation together with the grape skins. White wine is made by fermenting juice which is made by pressing crushed grapes to extract a juice; the skins are removed and play no further role. Occasionally white wine is made from red grapes, this is done by extracting their juice with minimal contact with the grapes’ skins. Rosé wines are made from red grapes where the juice is allowed to stay in contact with the dark skins long enough to pick up a pinkish color, but little of the tannins contained in the skins.
To start primary fermentation yeast is added to the must for red wine or juice for white wine. During this fermentation, which often takes between one and two weeks, the yeast converts most of the sugars in the grape juice into ethanol (alcohol) and carbon dioxide. The carbon dioxide is lost to the atmosphere. After the primary fermentation of red grapes the free run wine is pumped off into tanks and the skins are pressed to extract the remaining juice and wine, the press wine blended with the free run wine at the wine makers discretion. The wine is kept warm and the remaining sugars are converted into alcohol and carbon dioxide. The next process in the making of red wine is secondary fermentation. This is a bacterial fermentation which converts malic acid to lactic acid. This process decreases the acid in the wine and softens the taste of the wine. Red wine is sometimes transferred to oak barrels to mature for a period of weeks or months, this practice imparts oak aromas to the wine. The wine must be settled or clarified and adjustments made prior to filtration and bottling.
The time from harvest to drinking can vary from a few months for Beaujolais nouveau wines to over twenty years for top wines. However, only about 10% of all red and 5% of white wine will taste better after five years than it will after just one year. Depending on the quality of grape and the target wine style, some of these steps may be combined or omitted to achieve the particular goals of the winemaker. Many wines of comparable quality are produced using similar but distinctly different approaches to their production; quality is dictated by the attributes of the starting material and not necessarily the steps taken during vinification.
Variations on the above procedure exist. With sparkling wines such as Champagne, an additional fermentation takes place inside the bottle, trapping carbon dioxide and creating the characteristic bubbles. Sweet wines are made by ensuring that some residual sugar remains after fermentation is completed. This can be done by harvesting late (late harvest wine), freezing the grapes to concentrate the sugar (ice wine), or adding a substance to kill the remaining yeast before fermentation is completed; for example, high proof brandy is added when making port wine. In other cases the winemaker may choose to hold back some of the sweet grape juice and add it to the wine after the fermentation is done, a technique known as süssreserve.
The process produces wastewater, pomace, and lees that require collection, treatment, and disposal or beneficial use.

SINGLE CELL PROTEIN
Single cell protein (SCP) typically refers to sources of mixed protein extracted from pure or mixed cultures of algae, yeasts, fungi or bacteria (grown on agricultural wastes) used as a substitute for protein-rich foods, in human and animal feeds. 

History
Early history Since 2500 BC yeasts have been used in bread and beverage production. In 1781 processes for preparing highly concentrated forms of yeast were established. In 1919 Endomyces vernalis yielded fats from sulphite liquor (from paper manufacture), and similarly in 1941 employing Geotrichum.
“Food from oil”
In the 1960s, researchers at British Petroleum developed what they called “proteins-from-oil process”: a technology for producing single cell protein by yeast fed by waxy n-paraffins, a product produced by oil refineries. Initial research work was done by Alfred Champagnat at BP’s Lavera Oil Refinery in France; a small pilot plant there started operations in March in 1963, and the same construction of the second pilot plant, at Grangemouth Oil Refinery in Britain, was authorized. The term SCP was coined in 1966 by Carol L. Wilson at MIT.
The “food from oil” idea became quite popular by the 1970s, with Champagnat being awarded the UNESCO Science Prize in 1976, and paraffin-fed yeast facilities being built in a number of countries. The primary use of the product was as poultry and cattle feed.
The Soviets were particularly enthusiastic, opening large “BVK” (belkovo-vitaminny kontsentrat, i.e., “protein-vitamin concentrate”) plants next to their oil refineries in Kstovo (1973) and Kirishi (1974). The Soviet Ministry of Microbiological Industry had eight plants of this kind by 1989, when, pressured by the environmentalist movements, the government decided to close them down, or convert to some other microbiological processes.
SCP Production Process
Single cell proteins develop when microbes ferment waste materials (including wood, straw, cannery and food processing wastes, residues from alcohol production, hydrocarbons, or human and animal excreta). The problem with extracting single cell proteins from the wastes is the dilution and cost. They are found in very low concentrations, usually less than 5%. Engineers have developed ways to increase the concentrations including centrifugation, flotation, precipitation, coagulation and filtration, or the use of semi-permeable membranes.
The single cell protein needs to be dehydrated to approximately 10% moisture content and/or acidified to aid in storage and prevent spoilage. The methods to increase the concentrations to adequate levels, and de-watering process require equipment that is expensive and not always suitable for small-scale operations. It is economically prudent to feed the product locally and shortly after it is produced.

Examples:
Microbes employed include yeasts (Saccharomyces cerevisiae, Candida utilis=Torulopsis and Geotrichum candidum(=Oidium lactis)), other fungi (Aspergillus oryzae, Sclerotium rolfsii, Polyporus and Trichoderma), bacteria (Rhodopseudomonas capsulata). and algae (Chlorella and Spirulina). Typical yields of 43 to 56%, with protein contents of 44% to 60%.
The fungus Scytalidium acidophilum grows at below pH 1, offering advantages of:
- low-cost aseptic conditions,
- avoiding over 100-fold dilution of the acidic hydrolysates to pH values needed for other microbes
- after the biomass is harvested, the acids can be reused.
Commercial production of SCP (Spirulina) includes Cyanotech in Hawaii and Earthrise in California. TOPRINA- scp made from condidor lipolytica in uk PRUTEEN-SCP made from methanol. TORUTEIN- SCP made from ethanol using torula yeast.
Advantages of Production of SCP
Large-scale production of microbial biomass has many advantages over the traditional methods for producing proteins for food or feed.
1. Microorganisms have a high rate of multiplication to hence rapid succession of generation (algae: 2-6 hours, yeast: 1-3 hours, bacteria: 0.5-2 hours)
2. They can be easily genetically modified for varying the amino acid composition.
3. A very high protein content 43-85 % in the dry mass.
4. They can utilize a broad spectrum of raw materials as carbon sources, which include even waste products. Thus they help in the removal of pollutants also.
5. Strains with high yield and good composition can be selected or produce relatively easily.
6. Microbial biomass production occurs in continuous cultures and the quality is consistent since the growth is independent of seasonal and climatic variations.
7. Land requirements is low and is ecologically beneficial.
8. A high solar energy conversion efficiency per unit area.
9. Solar energy conversion efficiency can be maximized and yield can be enhanced by easy regulation of physical and nutritional factors.
10. Algal culture can be done in space which is normally unused and so there is no need to compete for land.
Microbes in Cheese and Yoghurt
Milk Protein Consists of proteins, lipids, lactose, minerals, vitamins and Enzymes such as oxidases, phosphatases, peroxidases, catalases, amylases and lipases.Casein makes up 80% of the milk proteincasein is precipitates along with other components when acidified. Milk clotting is done with rennet (chymosin) Rennet hydrolyses the bond between phenylalanine and methionine.
| CHEESE TYPE | EXAMPLE |
| Soft cheese Semi soft cheese Semi Hard Hard Very hard | Cambridge, Bondon Limburger, Brie Edam, Gouda Cheddar, Cheshire Parmesan, Romano |
NORMAL FLORA OF CHEESE MILK
- Corynebacteria
- Micrococci
- Enterococci
- Spores of Bacillus and Clostridium
- Staphylococci
- Coliforms
- Lactic acid bacteria
- Lactobacilli
- Pediococci
- Leuconostocs


L. casei L.acidophilus


Streptococcus salivaricus L.delbrueckii
PROCESS OF CHEESEMAKING: –

Yoghurt or yogurt is a dairy product produced by bacterial fermentation of milk. Fermentation of lactose produces lactic acid, which acts on milk protein to give yoghurt its texture and its characteristic tang. Dairy yoghurt is produced using a culture of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus salivarius subsp. thermophilus bacteria. The milk is heated to about 80 °C to kill any undesirable bacteria and to change the milk proteins so that they set together rather than form curds. It is then cooled to about 45 °C. The bacteria culture is added, and this temperature is maintained for 4 to 7 hours for fermentation. Soy yoghurt, a non-dairy yoghurt alternative, is made from soy milk.
People have been making and eating yogurt for at least 5,400 years. Today, it is a common food item throughout the world. A nutritious food with unique health benefits, it is rich in protein, calcium, riboflavin, vitamin B6 and vitamin B12.[1]
History
There is evidence of precultured milk products being produced as food for at least 4,500 years. The earliest yoghurts were probably spontaneously fermented by wild bacteria.
The oldest writings mentioning yogurt are attributed to Pliny the Elder, who remarked that certain nomadic tribes knew how “to thicken the milk into a substance with an agreeable acidity”.[7] The use of yoghurt by medieval Turks is recorded in the books Diwan Lughat al-Turk by Mahmud Kashgari and Kutadgu Bilig by Yusuf Has Hajib written in the 11th century.[8][9] Both texts mention the word “yoghurt” in different sections and describe its use by nomadic Turks.[8][9] An early account of a European encounter with yoghurt occurs in French clinical history: Francis I suffered from a severe diarrhoea which no French doctor could cure. His ally Suleiman the Magnificent sent a doctor, who allegedly cured the patient with yoghurt.[10][11] Being grateful, the French king spread around the information about the food which had cured him.
![]()
Raita is a condiment made with yoghurt and popular in India and Pakistan. Until the 1900s, yoghurt was a staple in diets of people in the Russian Empire (and especially Central Asia and the Caucasus), Western Asia, South Eastern Europe/Balkans, Central Europe, and India. Stamen Grigorov (1878–1945), a Bulgarian student of medicine in Geneva, first examined the microflora of the Bulgarian yoghurt. In 1905 he described it as consisting of a spherical and a rod-like lactic acid bacteria. In 1907 the rod-like bacteria was called Lactobacillus bulgaricus (now Lactobacillus delbrueckii subsp. bulgaricus). The Russian Nobel laureate biologist Ilya Ilyich Mechnikov, from the Institut Pasteur in Paris, was influenced by Grigorov’s work and hypothesised that regular consumption of yoghurt was responsible for the unusually long lifespans of Bulgarian peasants. Believing Lactobacillus to be essential for good health, Mechnikov worked to popularise yoghurt as a foodstuff throughout Europe.
Isaac Carasso industrialized the production of yoghurt. In 1919, Carasso, who was from Ottoman Salonika, started a small yoghurt business in Barcelona and named the business Danone (“little Daniel”) after his son. The brand later expanded to the United States under an Americanised version of the name: Dannon.
![]()
Tarator is a cold, refreshing soup made of yoghurt and cucumber (dill, garlic, walnuts and sunflower oil are sometimes added), popular in Bulgaria. Yoghurt with added fruit jam was patented in 1933 by the Radlická Mlékárna dairy in Prague. It was introduced to the United States in 1947, by Dannon.
Yoghurt was first introduced to the United States by Armenian immigrants Sarkis and Rose Colombosian, who started “Colombo and Sons Creamery” in Andover, Massachusetts in 1929.[13][14] Colombo Yogurt was originally delivered around New England in a horse-drawn wagon inscribed with the Armenian word “madzoon” which was later changed to “yogurt”, the Turkish name of the product, as Turkish was the lingua franca between immigrants of the various Near Eastern ethnicities[citation needed] who were the main consumers at that time. Yoghurt’s popularity in the United States was enhanced in the 1950s and 1960s, when it was presented as a health food. By the late 20th century yoghurt had become a common American food item and Colombo Yogurt was sold in 1993 to General Mills, which discontinued the brand in 2010.[15]
Nutritional value and health benefits
![]()
Tzatziki is an appetiser made with yoghurt, popular in Greece and Bulgaria, where it is called Dry Tarator. Yoghurt is nutritionally rich in protein, calcium, riboflavin, vitamin B6 and vitamin B12. It has nutritional benefits beyond those of milk. People who are moderately lactose-intolerant can consume yoghurt without ill effects, because much of the lactose in the milk precursor is converted to lactic acid by the bacterial culture. Yoghurt may also be used in preventing antibiotic-associated diarrhea. Yoghurt is believed to promote good gum health, possibly because of the effect of lactic acid present in yoghurt.
A study published in the International Journal of Obesity (11 January 2005) also found that the consumption of low-fat yoghurt can promote weight loss, especially due to the calcium in the yoghurt.
Varieties and presentation

![]()
Dadiah sold in Bukittinggi Market Dadiah, or Dadih, is a traditional West Sumatran yoghurt made from water buffalo milk. It is fermented in bamboo tubes. Yoghurt is popular in Nepal, where it is served both as an appetizer and dessert. Locally called dahi, it is a part of the Nepali culture, used in local festivals, marriage ceremonies, parties, religious occasions, family gatherings, and so on. The most famous type of Nepalese yoghurt is called juju dhau, originating from the city of Bhaktapur.
Tarator and Cacık are popular cold soups made from yoghurt, popular during summertime in Albania, Bulgaria, Republic of Macedonia, and Turkey. They are made with ayran, cucumbers, dill, salt, olive oil, and optionally garlic and ground walnuts. Tzatziki, a thick yoghurt-based sauce similar in concoction to tarator, is popular in Greece. Bulgaria typically calls tzatziki “dry tarator”.
Khyar w Laban (cucumber and yogurt salad) is a popular dish in Lebanon. Also, a wide variety of local Lebanese dishes are cooked with yogurt like “Kibbi bi Laban” etc..
Rahmjoghurt, a creamy yoghurt with much higher fat content (10%) than most yoghurts offered in English-speaking countries (Rahm is German for “cream”), is available in Germany and other countries.
Cream-top yoghurt is yoghurt made with unhomogenized milk. A layer of cream rises to the top, forming a rich yoghurt cream. Cream-top yoghurt was first made commercially popular in the United States by Brown Cow of Newfield, New York, bucking the trend toward low- and non-fat yoghurts. Jameed is yoghurt which is salted and dried to preserve it. It is popular in Jordan. Zabadi is the type of yoghurt made in Egypt, usually from the milk of the Egyptian water buffalo. It is particularly associated with Ramadan fasting, as it is thought to prevent thirst during all-day fasting.
Raita is a yoghurt-based South Asian/Indian condiment, used as a side dish. The yoghurt is seasoned with cilantro (coriander), cumin, mint, cayenne pepper, and other herbs and spices. Vegetables such as cucumber and onions are mixed in, and the mixture is served chilled. Raita has a cooling effect on the palate which makes it a good foil for spicy Indian dishes. Dudh is a Sindhi-curd, popular in India. People drink dudh along with food at intervals, to help digestion and make food more delicious. In some places dudh is also served with plain rice. Dahi is a yoghurt of the Indian subcontinent, known for its characteristic taste and consistency. The word dahi seems to be derived from the Sanskrit word dadhi, one of the five elixirs, or panchamrita, often used in Hindu ritual. Dahi also holds cultural symbolism in many homes in the Mithilanchal region of Bihar. It is found in different flavours, two of which are famous: sour yoghurt (tauk doi) and sweet yoghurt (meesti or podi doi). In India, it is often used in cosmetics mixed with turmeric and honey. Sour yoghurt is also used as a hair conditioner by women in many parts of India.
Srikhand, a popular dessert in India, is made from drained yoghurt, saffron, cardamom, nutmeg and sugar and sometimes fruits such as mango or pineapple.
Sweetened and flavored yoghurt
To offset its natural sourness, yoghurt can be sold sweetened, flavored or in containers with fruit or fruit jam on the bottom.[21] If the fruit has been stirred into the yoghurt before purchase, it is commonly referred to as Swiss-style.[22] Most yoghurts in North America] have added pectin, found naturally in fruit, and/or gelatin to artificially create thickness and creaminess at lower cost. This type of adulterated product is also marketed under the name Swiss-style, although it is unrelated to the way yoghurt is eaten in Switzerland. Some yoghurts, often called “cream line,” are made with whole milk which has not been homogenized so the cream rises to the top. Fruit jam is used instead of raw fruit pieces in fruit yoghurts to allow storage for weeks. Sweeteners such as cane sugar or sucralose – for low-calorie yogurts – are often present in large amounts in commercial yoghurt. In the USA, sweetened, flavored yoghurt is the most popular type, typically sold in single-serving plastic cups. Typical flavors are vanilla, honey, or fruit such as strawberry, blueberry, blackberry, raspberry, or peach.
Strained yoghurts
Strained yoghurts are types of yoghurt which are strained through a paper or cloth filter, traditionally made of muslin, to remove the whey, giving a much thicker consistency and a distinctive, slightly tangy taste.
Labneh is a strained yoghurt used for sandwiches popular in Arab countries. Olive oil, cucumber slices, olives, and various green herbs may be added. It can be thickened further and rolled into balls, preserved in olive oil, and fermented for a few more weeks. It is sometimes used with onions, meat, and nuts as a stuffing for a variety of pies or kebbeh balls. Some types of strained yoghurts are boiled in open vats first, so that the liquid content is reduced. The popular East Indian dessert, a variation of traditional dahi called mishti dahi, offers a thicker, more custard-like consistency, and is usually sweeter than western yoghurts. Strained yoghurt is also enjoyed in Greece and is the main component of tzadziki, a well-known accompaniment to gyros and souvlaki pita sandwiches.
YOGURT PRODUCTS:


Fruit-on-the-bottom style Soft-serve and Hard Pack frozen yogurt
| Download this lecture as PDF here |

 with cork lids, and show that microbes would soon arise.
with cork lids, and show that microbes would soon arise.